Abstract
We sequenced the complete mitochondrial (mt) genome of Chloris sinica ussuriensis. The circular mt genome is 16,813 bp long and encodes 13 proteins, 22 transfer RNAs, and 2 ribosomal RNAs. Phylogenetic analysis based on full mt genome sequences confirmed that the C. s. ussuriensis is monophyletic group of the Chloris sinica. The complete mitochondrial genome of C. s. ussuriensis can provide a valuable data for resolving geographical distribution of evolutionary subdivision within the C. sinica species in East Asia.
The Grey-capped Greenfinch Chloris sinica (Aves: Passeriformes: Fringillidae) is a small passerine bird in the Fringillidae family, which has limited ranges of distribution in East Asia. This species is now regarded as consisting of five to six subspecies (del Hoyo et al. Citation2010; Gill and Donsker Citation2014), but the phylogenetic relationships are still unclear and additional taxon sampling and sequence data are needed. Here, we present the complete mitochondrial (mt) genome of C. s. ussuriensis to help elucidate the phylogeographic patterns of the C. sinica species.
During the breeding season, the C. s. ussuriensis specimen was collected from Taean, Chungcheong-do, Korea. We extracted the genomic DNA from the blood sample using the DNeasy Blood & Tissue kit (Qiagen, Valencia, CA) as described by the manufacturer’s protocol, and determined the complete mt genome sequence using the primer-walking approach. The blood sample used in this study was deposited at the Wildlife Specimen Bank in Chonnam National University, Korea.
The complete mt genome of C. s. ussuriensis was 16,813 bp in length (GenBank accession No. MH047559), and consisted of 13 protein-coding genes, 22 transfer RNA (tRNA) genes, 2 ribosomal RNA genes (rRNAs), an origin of light strand replication site (OL), and a putative long noncoding region called the control (D-loop) region, agreeing with the typical vertebrate gene arrangement. Most of the mt genes were encoded on the H-strand, with the exception of the one protein-coding gene (nad6) and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAPro, and tRNAGlu) that were encoded on the L-strand. The overall nucleotide composition for C. s. ussuriensis was 30.7% A, 30.5% C, 14.1% G, and 24.7% T.
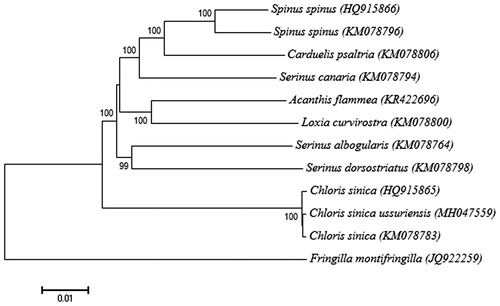
The phylogenetic analysis of full mt genome of C. s. ussuriensis with two available complete mt genomes from Chloris genus and nine ecologically close species revealed that C. s. ussuriensis was the most closely related to the Chloris sinica (). The present study provides essential mt genome data for further intraspecific phylogeography and population differentiation among the Grey-capped Greenfinches in East Asia.
Figure 1. Phylogeny of Chloris sinica ussuriensis and other related species with Fringilla montifringilla as outgroup. The phylogenetic tree derived from complete mitochondrial (mt) genome sequences was constructed by a neighbour-joining method with 1000 bootstrap replicates in the program MEGA7 (Saito and Nei 1987). GenBank accession numbers of each mt genome sequence are given in the bracket after the species name. In accordance with the recently proposed nomenclature (Zuccon et al. Citation2012), in this study, Carduelis sinica was named as Chloris sinica.
GenBank accession number from the complete mitochondrial genome of Chloris sinica ussuriensis (MH047559) has been registered with the NCBI database.
Disclosure statement
The authors declare no conflict of interests.
Additional information
Funding
References
- del Hoyo J, Elliott A, Christie DA. 2010. Handbook of the Birds of the world. Vol. 15. Barcelona: Lynx Editions.
- Gill F, Donsker D. 2014. IOC World Bird List Version 4.2. [cited 2014 Apr 1]. www.worldbirdnames.org.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Zuccon D, Prŷs-Jones R, Rasmussen PC, Ericson PGP. 2012. The phylogenetic relationships and generic limits of finches (Fringillidae). Mol Phylogenet Evol. 62:581–596.
