Abstract
The first complete mitochondrial genome of a Kaloula verrucosa frog was characterized in this work. The mitogenome was 17,061 base pairs (bp) in length, containing 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes, and a control region (D-loop). The overall base composition was 29.65% A, 30.77% T, 25.41% C and 14.17% G. Besides, the gene arrangement was identical to that observed in vertebrates. Five of 13 PCGs (COII, ATP6, COIII, ND3 and ND4) were ended with incomplete stop codon T. Except for ND6 gene encoded on L-strand, all other PCGs were encoded on H-strand. The non-coding region was 1665 bp in size, which was heavily biased to A + T (65.77%). Additionally, we found mitogenome size of all sequenced Kaloula species were bigger than that of Microhyla species, which was ascribe to the difference of D-loop size. Phylogenetic analysis showed that K. verrucosa was the sister species of Kaloula regifera. This work will provide basic molecular data for further molecular evolution and phylogenetic research of K. verrucosa and other microhylids.
Kaloula Gray 1831 is a genus of 17 species, showing a broad distributional range in Korea and northern China to Lesser Sundas and Philippines, Bangladesh, and India, controversially in Nepal (Frost Citation2018). However, only three species (Kaloula pulchra AY458595, Kaloula borealis NC_020044, K. rugifera KP682314) of this genus were determined the complete mtDNA sequence (Hwang and Lee Citation2012; Wu et al. Citation2014; Deng et al. Citation2016). Therefore, more complete mitochondrial genome species would improve our understanding for further genetic research and conservation of this genus. Kaloula verrucosa Boulenger, 1904 is an endemic species of China, occurring in Sichuan, Yunnan, and Guizhou (Yang and Wu Citation2004; Jiang et al. Citation2016; Jiang et al. Citation2016). Here, Genomic DNA extraction, mtDNA amplification and sequencing were from leg muscle preserved in 95% ethanol of a single K. verrucosa specimen collected from Huili, Sichuan (26.74198°N and 102.48511°E) and the specimen was stored in department of Herpetology of Chengdu Institute Biology (number 20090351). The amplified primers were used as described by Zhang et al. (Citation2013) and amplified methods were performed according to Khatiwada et al. Citation2017.
The total length of complete mitogenome was 17,061 bp (GenBank accession no. MG962359), which included 13 PCGs, two ribosomal RNA genes, 22 transfer RNA (tRNA) genes, and a control region (D-loop). The gene organization was similar to that observed in most microhylids (Wu et al. Citation2014; Deng et al. Citation2016; Wang et al. Citation2016). The overall base composition was 29.65% A, 30.77% T, 25.41% C, and 14.17% G. The AT content (60.42%) was much higher than that of GC (39.58%). ATP8 gene (165 bp) was the shortest while ND5 was the longest (1806 bp) among the 13 PCGs. Except for ND6 and 8 tRNA genes (tRNA-Pro, tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, and tRNA-Glu) encoded on L-strand, the remaining 28 genes were encoded on H-strand. Most of the PCGs were terminated with a complete stop codon (TAG, AGG, TAA or AGA), however, five genes (COII, ATP6, COIII, ND3 and ND4) were stopped with incomplete T. Each typical tRNA cloverleaf secondary structure is predicted by tRNA-Scan Web Server (Lowe and Chan Citation2016). Total 22 tRNA genes, with the size ranging from 57 bp to 73 bp, were interspersed along the whole genome, and most of the tRNAs could form a cloverleaf structure. The putative origin of L strand replication (OL), with size of 29 bp between the tRNA-Asn and tRNA-Cys genes, could be folded into a stem loop of secondary structure, which was similar to that of other vertebrates (Deng et al. Citation2016). The D-loop region (1665 bp), located between Cytb and tRNA-Leu, was heavily biased to A + T (65.77%). In addition, the mitogenome sizes of sequenced Microhyla species (Mean ± SD, 16,729.29 ± 19.49 bp) were smaller than that of Kaloula species (17,031.5 ± 151.09 bp), which might be ascribe to the difference of D-loop size (Microhyla: 1341.29 ± 18.06; Kaloula: 1629.75 ± 152.48). D-loop region which was the most variable part may play a key role in molecular evolution and phylogenetic research.
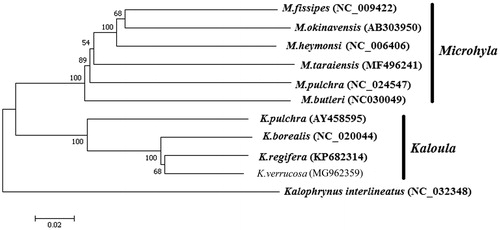
Neighbour-joining algorithm tree was constructed based on 11 complete mtDNA sequences of available Microhylidae (). The result showed that Microhyla, and Kaloula were from two strong monophyletic clades and K. verrucosa is the sister species of K. regifera.
Figure 1. Neighbour-joining algorithm tree was constructed based on 11 complete mitochondrial genome sequences. The branches were validated by bootstrap analysis from 1000 replications and the numbers in branch nodes were bootstrap support values. The position of K. verrucosa was not shown in bold and Kalophrynus interlineatus was used as outgroup.
Acknowledgements
We are thankful to Jiongyu Liu for collecting specimens.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Deng XY, Wang SH, Liang XX, Jiang JP, Wang B, Deng LJ. 2016. The complete mitochondrial genome of Kaloula rugifera (Amphibia, Anura, Microhylidae). Mitochondrial DNA. 27:3391–3392.
- Frost DR. 2018. Amphibian Species of the World: an Online Reference. Version 6.0, vol. 2018. American Museum of Natural History. New York, USA. [accessed 2018 April 10]. http://research.amnh.org/herpetology/amphibia/index.html.
- Jiang JP, Xie F, Zang CX, Cai L, Li C, Wang B, Li JT, Wang J, Hu JH, Wang Y, Liu JY. 2016. Assessing the threat status of amphibians in China. Biodivers Sci. 24:588–597.
- Jiang ZG, Jiang JP, Wang YZ, Zhang E, Zhang YY, Li LL, Xie F, Cai B, Cao L, Zheng GM, et al. 2016. Red List of China’s Vertebrates. Biodivers Sci. 24:500–551.
- Hwang DS, Lee JS. 2012. Complete mitochondrial genome of the boreal digging frog Kaloula borealis (Anura, Microhylidae). Mitochondrial DNA. 23:301–302.
- Khatiwada JR, Wang SH, Shu GC, Xie F, Jiang JP. 2017. The mitochondrial genome of the Microhyla taraiensis (Anura: Microhylidae) and related phylogenetic analyses. Conserv Genet Resour. 2017:1–4.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Wang SH, Liu LS, Jiang JP. 2016. The complete mitochondrial genome of Microhyla butleri (Amphibia, Anura, Microhylidae). Mitochondrial DNA Part B. 1:154–155.
- Wu XY, Li YM, Zhang HB, Yan L, Wu XB. 2014. The complete mitochondrial genome of Microhyla pulchra (Amphidia, Anura, Microhylidae). Mitochondrial DNA. 27:40–41.
- Yang DT, Wu GF. 2004. Kaloula verrucosa. The IUCN Red List of Threatened Species 2004:e.T57859A11695288. [accessed 2018 April 10]. http://dx.doi.org/10.2305/IUCN.UK.2004.RLTS.T57859A11695288.en.
- Zhang P, Liang D, Mao R-L, Hillis DM, Wake DB, Cannatella DC. 2013. Efficient sequencing of anuran mtDNAs and a mitogenomic exploration of the phylogeny and evolution of frogs. Mol Biol Evol. 30:1899–1915.
