Abstract
A complete mitochondrial genome sequence was determined for a member of the Caridina indistincta species complex known as C. indistincta ‘sp. A’. The 15,461 bp sequence (GenBank: MH189850) was obtained via genome skimming, and contains 13 protein coding genes, 22 tRNA genes, 2 rRNA genes, and a 646 bp control region arranged in the pancrustacean ground pattern. Caridina indistincta sp. A is a freshwater macroinvertebrate important for ecosystem health monitoring in Australia and this reference will be a useful resource for metabarcoding and eDNA studies.
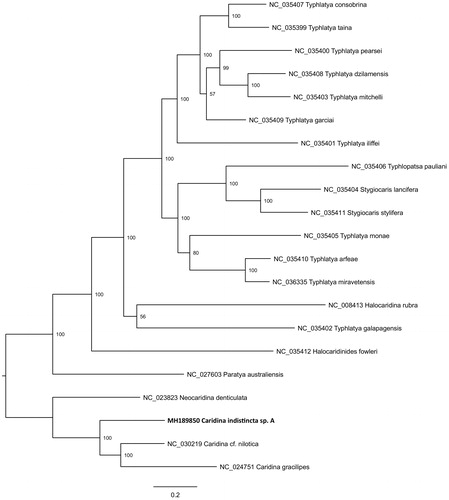
Caridina indistincta is a species complex of small freshwater atyid shrimps, widely distributed in eastern Australia (Page et al. Citation2005). The complex appears to consist of at least five species, informally known as C. indistincta sp. A, sp. B, sp. C, sp. D, and sp. E which can be morphologically diagnosed and are genetically divergent based on mtDNA (Page et al. Citation2005). The taxon C. indistincta sp. A is geographically restricted to 11 coastal basins spanning ∼350 km in southeastern Queensland, Australia, and is an indicator of biological condition in ecosystem health monitoring programs for this region (Page and Hughes Citation2007). A shallow whole-genome shotgun library of C. indistincta sp. A was prepared from a specimen collected in Mimosa Creek (27.54973 S, 153.05188 E) located on the Nathan Campus of Griffith University, Australia. Details of library preparation and sequencing (Illumina Miseq, insert size ∼500 bp) are reported elsewhere (Schmidt, Brockett, et al. Citation2016; Schmidt, Islam, et al. Citation2016). A total of 5.8 × 106 reads were generated. Novoplasty v2.6.5 was used to assemble 3202 reads into a circular contig of 15,461 bp length with average coverage of ×82 (Dierckxsens et al. Citation2017). A seed sequence used to initiate assembly was retrieved using the mitogenome of Caridina gracilipes (NC_024751.1; Xu et al. Citation2016). The Mitos WebServer (Bernt et al. Citation2013) was used for initial gene annotation, followed by manual adjustment of gene boundaries after alignment with existing Caridina reference genomes (i.e. GenBank: KU726823; KM023648). The fully annotated new mitogenome is available at GenBank accession MH189850. No anomalies were detected – 13 protein coding genes, 22 tRNA genes, two rRNA genes and a 646 bp control region were arranged in the pancrustacean ground pattern which is common to other members of the Atyidae characterized to date (Tan et al. Citation2017). In a phylogenetic context, MH189850 was analysed with all available atyid mitogenomes on the NCBI RefSeq database, demonstrating its placement with other Caridina spp (). A BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) showed 100% identity between the new mitogenome and a 450 bp fragment of COI identified as C. indistincta sp. A, voucher GU84 sampled from the South Pine River, which is geographically proximate to the sampling location of the specimen analysed here (GenBank: JX913903; Page and Hughes Citation2014).
Figure 1. Phylogenetic placement of the new Caridina indistincta sp. A mitogenome (GenBank: MH189850) relative to 20 Atyidae mitogenomes sourced from the NCBI RefSeq database. Tip labels include GenBank accession and species name; node labels show bootstrap result. Alignment of mitogenomes (excluding 16S, 12S, and control region) was performed using MAFFT v7.017 (Katoh et al. Citation2002). A maximum likelihood phylogenetic analysis was performed on the final alignment of 12 821 bp with RAxML v8.2.11 using the GTR + GAMMA substitution model with 1000 bootstrap replicates (Stamatakis Citation2006).
Acknowledgements
The sample used for sequencing was collected by Amaal Ghazi Yasser (Griffith University) and the DNA extraction was performed by Kathryn Real (Griffith University). Assistance with the Illumina MiSeq sequencing run was provided by Nicole Hogg and Fraxa Caraiani at Griffith University DNA Sequencing Facility.
Disclosure statement
The author reports no conflicts of interest. The author alone is responsible for the content and writing of the paper.
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Page TJ, Choy SC, Hughes JM. 2005. The taxonomic feedback loop: symbiosis of morphology and molecules. Biol Lett. 1:139–142.
- Page TJ, Hughes JM. 2007. Radically different scales of phylogeographic structuring within cryptic species of freshwater shrimp (Atyidae: Caridina). Limnol Oceanogr. 52:1055–1066.
- Page TJ, Hughes JM. 2014. Contrasting insights provided by single and multispecies data in a regional comparative phylogeographic study. Biol J Linn Soc. 111:554–569.
- Schmidt DJ, Brockett B, Espinoza T, Connell M, Hughes JM. 2016. Complete mitochondrial genome of the Endangered Mary River turtle (Elusor macrurus) and low mtDNA variation across the species’ range. Aust J Zool. 64:117–121.
- Schmidt DJ, Islam MRU, Hughes JM. 2016. Complete mitogenomes for two lineages of the Australian smelt, Retropinna semoni (Osmeriformes: Retropinnidae). Mitochondrial DNA B Resour. 1:615–616.
- Stamatakis A. 2006. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Tan MH, Gan HM, Lee YP, Poore GCB, Austin CM. 2017. Digging deeper: new gene order rearrangements and distinct patterns of codons usage in mitochondrial genomes among shrimps from the Axiidea, Gebiidea and Caridea (Crustacea: Decapoda). PeerJ. 5:e2982.
- Xu GC, Du FK, Nie ZJ, Xu P, Gu RB. 2016. Complete mitochondrial genome of Caridina nilotica gracilipes. Mitochondrial DNA Part A. 27:1249–1250.
