Abstract
Triaenophora shennongjiaensis (Orobanchaceae sensu lato) is a recently described rare and endangered species endemic to Central China. In this study, the complete chloroplast (cp) genome of T. shennpongjiaensis was assembled based on reads obtained with the Illumina HiSeq platform. The cp genome of T. shennongjiaensis was 15,5319 bp in length and contained a pair of inverted repeat (IR, 27,484 bp) regions separated by a small single copy (SSC, 15,450 bp) and a large single copy (LSC, 84,901 bp) region. It encoded 112 genes including 78 protein-coding genes, 30 tRNA genes, and eight ribosomal RNA genes. The overall AT content of T. shennongjiaensis cp genome is 61.9%. The maximum likelihood phylogenetic analysis supports T. shennongjiaensis as sister to Rehmannia. This result will be helpful for the systematics, conservation, and breeding programs of Triaenophora.
Introduction
Triaenophora is a small endemic genus in Central China consisting of three narrowly distributed species formerly placed in Scrophulariaceae (Hong et al. Citation1998; Li et al. Citation2005). Triaenophora shennongjiaensis is an endangered species from the Shennongjia National Natural Reserve, Hubei, China (Li et al. Citation2005). Recent molecular systematic studies showed that Triaenophora is not part of Scrophulariaceae and have placed Triaenophora in Orobanchaceae s.l. (Albach et al. Citation2009; Xia et al. Citation2009). In this study, we report the complete chloroplast (cp) genome of T. shennongjiaensis.
The plant material of T. shennongjiaensis was sampled from Panlong Cavern, Shennongjia National Natural Reserve, Hubei, China. The voucher specimen (ZX-2017-0601) is kept at the Henan Agricultural University Herbarium (HEAC). Genomic DNA was extracted from leaf tissue using the Plant Genomic DNA Kit (DP305) from Tiangen Biotech (Beijing) Co., Ltd. (Beijing, China). DNA sample was randomly fragmented into 400–600 bp fragments using an ultrasonicator. An Illumina paired-end DNA library with 500-bp insert size was constructed using a NEBNext® UltraTM DNA Library Prep Kit. Paired-end sequencing (2 × 150 bp) was conducted on an Illumina HiSeq × Ten platform.
The paired-end reads were qualitatively assessed and assembled with SPAdes 3.6.1 (Bankevich et al. Citation2012) using the Rehmannia piasezkii chloroplast genome sequence as a reference (GenBank accession KX636160) (Zeng et al. Citation2017). Small gaps in the assemblies were bridged with specific primers designed for PCR based on their flanking sequences and then by Sanger sequencing (Dong et al. Citation2013). Chloroplast genome annotation was performed with Plann (Huang and Cronk Citation2015). The cp genome sequence was submitted to GenBank (accession number MH071405).
The cp genome of T. shennongjiaensis was 155,319 bp in length and contains a pair of inverted repeat (IRa and IRb) regions of 27,484 bp, the large single copy (LSC) region and small single copy (SSC) region with the lengths of 84,901 and 15,450 bp, respectively (). The whole chloroplast genome encoded 112 genes including 78 protein-coding genes (PCG), 30 tRNAs, and four rRNA operons. Among these genes, 15 genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rps12, rpl16, trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) harboured one intron and two genes (clpP and ycf3) had two introns. Most genes occurred in a single copy, however, eight PCG genes (ndhB, rpl2, rpl23, rps7, rps12, ycf1, ycf2, and ycf15), seven tRNA genes (trnA-UGC, trnH-CAU, trnI-GAU, trnLCAA, trnN-GUU, trnR-ACG, and trnV-GAC), and four rRNA genes (rrn4.5, rrn5, rrn16, and rrn23) in the IR regions were duplicated. The overall AT content of the T. shennongjiaensis chloroplast genome is 61.9% and the corresponding values in LSC, SSC, and IR regions are 64.2, 68.1, and 57.5%, respectively.
Figure 1. Maximum likelihood phylogenetic tree based on 24 complete chloroplast genome sequences. Accession numbers: Cistanche deserticola KC128846, Cistanche phelypaea HG515538, Orobanche californica HG515539, Schwalbea americana HG738866, Lindenbergia philippensis HG530133, Rehmannia chingii KX426347, R. glutinosa KX636157, Rehmannia elata KX636161, Rehmannia piasezkii KX636160, Rehmannia solanifolia KX636159, Rehmannia henryi KX636158, Seasamum indicum JN637766, Scrophularia takesimensis KM590983, Premna microphylla KM981744, Tectona grandis HF567869, Scutellaria baicalensis KR233163, Origanum vulgare JX880022, Salvia miltiorrhiza JX312195, Salvia rosmarinus KR232566, Andrographis paniculate KF150644, Utricularia gibba KC997777, Boea hygrometrica JN107811, Olea europaea GU931818 and Triaenophora shennongjiaensis MH071405. The number on each node indicates the bootstrap value.
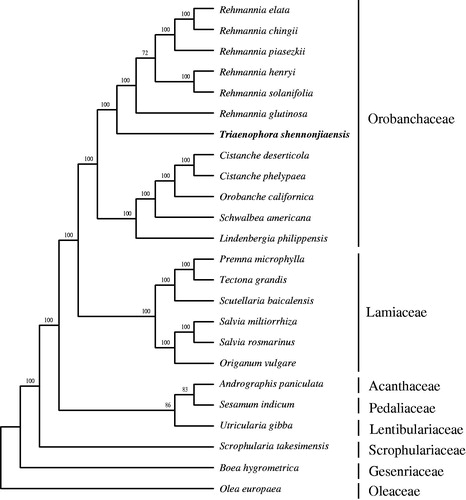
Plastome sequences of 24 Lamiales species including T. shennongjiaensis were aligned with MAFFT (Katoh and Standley Citation2013). A maximum likelihood analysis was performed with the RAxML software (Stamatakis Citation2014) using 1000 bootstrap replicates. All sampled members of Orobanchaceae s.l. (Lindenbergia, Rehmannia, Triaenophora, Cistanche, Orobanche, and Schwalbea) formed a clade and T. shennongjiaensis was sister to Rehmannia (). The chloroplast resource may be utilized for DNA barcoding, conservation genetics, and breeding of Triaenophora.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Albach DC, Yan K, Jensen SR, Li HQ. 2009. Phylogenetic placement of Triaenophora (formerly Scrophulariaceae) with some implications for the phylogeny of Lamiales. Taxon. 58:749–756.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to aingle-cell sequencing. J Comput Biol.19:455–477.
- Dong W, Xu C, Cheng T, Lin K, Zhou S. 2013. Sequencing angiosperm plastid genomes made easy: a complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol Evol. 5:989–997.
- Hong DY, Yang HB, Jin CL, Holmgren NH. 1998. Scrophulariaceae. In: Flora of China Editorial Committee, editor. Flora of China, vol. 18. Beijing/St. Louis: Science Press/Missouri Botanical Garden Press; p. 1–212.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Li XD, Li JQ, Zan Y-Y. 2005. A new species of Triaenophora (Scrophulariaceae) from China. Novon. 15:559–561.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Xia Z, Wang YZ, Smith JF. 2009. Familial placement and relations of Rehmannia and Triaenophora (Scrophulariaceae s.l.) inferred from five gene regions. Am J Bot. 96:519–530.
- Zeng SY, Zhou T, Han K, Yang YC, Zhao JH, Liu ZL. 2017. The complete chloroplast genome sequences of six Rehmannia species. Genes. 8:103.
