Abstract
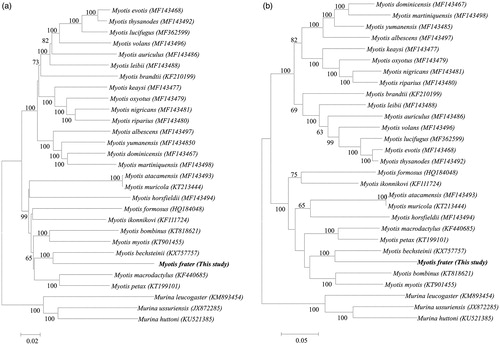
Here, we report the complete mitogenome of Myotis frater with the GenBank accession number MH177276 as a first step to elucidate genetic characteristics of this species. Its mitogenome was 17,089 bp long and consisted of 13 protein-coding genes (PCGs), two rRNA genes, 22 tRNA genes, and a control region. The gene order and composition of M. frater was similar to that of most other vertebrates. The base composition of the 13 PCG in descending order was A (33.8%), C (22.7%), T (30.4%), and G (13.1%), with an AT content of 64.2%. Four overlapping regions in ATP8/ATP6, ATP6/COX3, ND4L/ND4, and ND5/ND6, among the 13 PCGs were found. The 935 bp long control region is located between tRNA-Pro and tRNA-Phe with 4 ATTACATAATACATTATATGTATAATCGTACATTAAATTAACTCCCACATGAATATTAAGCATGTCCATACTAATATTAAT-repeat at 5′ region and 45 ACGCAT-repeat at 3′ terminus. Phylogenetic analysis suggested that M. frater is most closely related to M. bechsteinii (KX757757), it was supported by 100% bootstrap under both ML and NJ tree.
The Myotis frater has only been recorded twice in Korea, and their morphology and ecology are virtually unknown (Chung et al. Citation2017). We collected carcass of a female long-tailed whiskered bat in May 2016, in Gangwon Province, and to the best of our knowledge, this has been done for the first time in ∼30 years (Chung et al. Citation2017). Therefore, we report the complete mitogenome of M. frater as a first step to elucidate genetic characteristics of this species. The voucher specimen was deposited in the bat collection of the DGU-A004939 at the Dongguk University in Korea with the accession number of MH177276. Total genomic DNA was extracted from the wing membrane tissue using the DNeasy_Blood & Tissue Kit (Qiagen, Valencia, CA). We were used to design primers for PCR amplification and also used as templates for gene annotation from previously published complete mitogenome study (Yoon et al. Citation2013) and NCBI data base. PCR products were sequenced using the primer walking method using ABI 3730XL DNA Analyzer (Applied Biosystems Inc., Foster City, CA). The DNA sequences were assembled, aligned, and annotated using Genetyx 5.2 (Genetyx Corp., Tokyo, Japan) and tRNAscan-SE 1.21 (Lowe and Eddy Citation1997). The total length of the mitogenome of Myotis frater is 17,089 bp with a total base composition of 33.8% A, 30.4% T, 22.7% C and 13.1% G. It consisted of two rRNA genes, 22 tRNA genes, 13 protein-coding genes (PCGs), and one control region. Most genes were encoded on the H-strand with the exception of one PCG (ND6) and eight tRNA genes. The 12S rRNA and 16S rRNA were 969 bp and 1579 bp long, respectively. As in other vertebrate mitogenome, these were located between tRNA-Phe and tRNA-Leu-UUR, and separated by tRNA-Val. The complete mitogenome contained 22 tRNA genes which were interspersed along the genome with the size ranged from 60 to 75 bp. Most tRNAs could be folded into a cloverleaf secondary structure, with the exception of tRNA-Ser-AGY, which lacked the ‘DHU’ arm. The tRNA-Ser-AGY gene was identified by proposed secondary structures and the anti-codon (Kumazawa and Nishida Citation1993). Of the 13 PCGs, gene overlaps were observed between four pairs of the contiguous genes, i.e. ATP8–ATP6, ATP6–COX3, ND4L–ND4, and ND5–ND6, by 43, 1, 7 and 16 nucleotides, respectively. The majority of PCGs (10 of 13 genes) started with ATG, with three exception of ND2, ND3, and ND5, by starting with ATT, ATA, and ATA, respectively. Four types of stop codons including two complete codons, TAA or AGA, and two incomplete codons, T or TA were used in the PCGs. The 935 bp long control region is located between tRNA-Pro and tRNA-Phe with 4 ATTACATAATACATTATATGTATAATCGTACATTAAATTAACTCCCACATGAATATTAAGCATGTCCATACTAATATTAAT-repeat at 5′ region and 45 ACGCAT-repeat at 3′ terminus. Our phylogenetic analysis of the 13PCGs data sets also resulted in highly concordant branching topologies using neighbour-joining and maximum-likelihood methods. Phylogenetic analysis suggested that M. frater is most closely related to M. bechsteinii (KX757757), it was supported by 100% bootstrap under both ML and NJ tree ().
Figure 1. The phylogenetic relationship of M. frater and its allied 25 species were inferred from (a) neighbour-joining and (b) maximum likelihood methods analysis based on 13PCGs sequences with 1000 bootstrap replications. The bootstrap support, based on 1000 replicates, is indicated in each node with the 60% majority-rule tree values.
Acknowledgements
The authors thank anonymous reviewers for providing valuable comments on the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Chung CU, Kim SC, Jeon YS, Han SH. 2017. Morphological characteristics of long-tailed whiskered bat Myotis frater. J Environ Sci Int. 26:529–553.
- Kumazawa Y, Nishida M. 1993. Sequence evolution of mitochondrial tRNA genes and deep-branch animal phylogenetics. J Mol Evol. 37:380–398.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955.
- Yoon KB, Cho JY, Park YC. 2013. Complete mitochondrial genome of the Korean ikonnikov's bat Myotis ikonnikovi (Chiroptera: Vespertilionidae). Mitochondrial DNA. 26:274–275.
