Abstract
We report four mitochondrial genomes of South American electric knifefishes, derived from target capture and Illumina sequencing (HiSeq 2500 PE100). Two trans-Andean species Eigenmannia humboldtii (mitochondrial consensus genome of 25 individuals) and Sternopygus aequilabiatus (mitochondrial consensus genome of 30 individuals) from Colombia and two cis-Andean species Eigenmannia limbata from Suriname and Sternopygus macrurus from Argentina. Regarding Eigenmannia humboldtii, Eigenmannia limbata, and Sternopygus macrurus mitochondrial genomes have 13 protein-coding genes, 1 D-loop, 2 ribosomal RNAs, 22 transfer RNAs, and are 13,394 bp, 10,921 bp, and 13,013 bp in length respectively, for Sternopygus aequilabiatus mitochondrial genomes have 13 protein-coding genes, 2 ribosomal RNAs, 22 transfer RNAs, and is 14,270 bp in length.
Family Sternopygidae conform a group of species commonly known as glass knifefishes that are broadly distributed throughout, most of the, Neotropics on both sides of the Andes, from Panama to northern Argentina (Albert Citation2001, Citation2003). This family comprises 6 genera (Sternopygus, Eigenmannia, Rhabdolichops, Archolaemus, Distocyclus, and Japigny) and 41 species (Eschmeyer et al. Citation2018). This study reports the complete mitochondrial genome for four different species belonging to the order Gymnotiformes, family Sternopygidae that has not been sequenced so far and provides important information for future studies.
In this work, 25 individuals, in total, of Eigenmannia humboldtii were collected in field at Atrato, Cauca, Magdalena, San Jorge, and Tuira rivers basins and 30 individuals, in total, of Sternopygus aequilabiatus were collected in field at Atrato, Cauca, Magdalena, Rancheria, San Jorge, and Tuira rivers basins; Eigenmannia limbata specimen was collected in the field in Suriname river and a Sternopygus macrurus tissue was from Parana (Argentina) belonging to the STRI collection ; for sampling see Appendix S1 (Supporting information). Total DNA was obtained from 20 mg of muscle with the BioSprint 96 from QIAGEN® at The Smithsonian Tropical Research Institute (STRI). Following extraction, we quantified DNA extracts using a Qubit fluorometer (Life Technologies, Inc., Waltham, MA, USA) and samples were sent to MYcroarray, Inc (Ann Arbor, MI, USA) for a targeted sequencing approach. Probes were targeted (>200 base pairs) using target capture and Illumina sequencing (HiSeq 2500 PE100, Chicago, IL, USA). The capture target design is described in detail by Arcila et al. (Citation2017). Although probes were not designed to target mtDNA genes, we tested whether the raw Illumina data contained mtDNA genomes.
We mapped reads from each sample to its most relative available mitochondrial genome of reference (Eigenmannia sp. (Genbank AB054131) for Eigenmannia individuals and Sternopygus aequilabiatus (unpublished) for Sternopygus individuals) using SAMTOOLS (v1.3.1; Li et al. Citation2009). Samples were mapped to reference, trimmed and the consensus sequences were obtained using GENEIOUS (v8.1.9; Kearse et al. Citation2012). A consensus sequence per each species was extracted from the multiple alignment and posteriorly were cleaned and visually inspected to detect stop codons. The mtDNA genome was annotated using MitoAnnotator (Iwasaki at al. Citation2013).
The mitochondrial genomes of Eigenmannia humboldtii (Genbank MH263668), Eigenmannia limbata (Genbank MH263669), and Sternopygus macrurus (MH263671) are complete, with 13 protein-coding genes, 1 D-loop, 2 ribosomal RNAs, and 22 transfer RNAs for a total length of 13,394, 10,921, and 13,013 bp, respectively. For Sternopygus aequilabiatus (MH263670) we couldn’t recover D-loop, this genome has 13 protein-coding genes, 2 ribosomal RNAs, and 22 transfer RNAs for a total length of 14,270 bp. Base compositions are typical for vertebrate mitochondrial genomes with 27.4% A, 29.1% C, 18.0% G, and 25.4% T for E. humboldtii, 27.5% A, 28.3% C, 18.7% G, and 25.4% T for E. limbata, 29.7% A, 27.8% C, 15.6% G, and 26.9% T, for S. aequilabiatus and 29.7% A, 27.4% C, 16.0% G, and 26.8% T for S. macrurus.
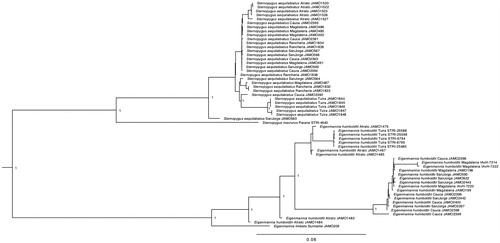
We reconstructed a phylogenetic tree based on the complete mitochondrial genome of 57 individuals belonging to the Sternopygidae family. The Bayesian tree was performed using MrBayes (v3.2.6; Huelsenbeck and Ronquist Citation2001; Ronquist and Huelsenbeck Citation2003) for 8 MCMC runs, 10 million of generations and sampled every 1000. Cis-andean species E. limbata and S. macrurus are both the sister group of the trans-Andean species; a remarkable find, is that individuals belonging to the great Magdalena basin falls in the same cluster ().
Figure 1. Molecular phylogeny of four South American electric knifefishes based on complete mitogenome. Support values at each node are Bayesian posterior probabilities. Branch label include information about sampled basin and tissue availability (JAMO: Pontificia Universidad Javeriana; IAvH: Instituto de Investigaciones Alexander von Humboldt; STRI: Smithsonian Tropical Research Institute).
AppendixS1.xlsx
Download MS Excel (12.8 KB)Acknowledgements
The authors would like to thank Instituto de Investigaciones Alexander von Humboldt (Colombia) and Smithsonian Tropical Research Institute (Panama) for helping with sample collection.
Disclosure statement
The authors do not report any conflict of interests, and are responsible for the content of this publication.
Additional information
Funding
References
- Albert JS. 2001. Species diversity and phylogenetic systematics of American knifefishes (Gymnotiformes, Teleostei). Ann Arbor: University of Michigan.
- Albert JS. 2003. Check list of the freshwater fishes of South and Central America family. In: Reis RE, Kullander SO, Ferraris CJ Jr, organizers, editors. Sternopygidae. Porto Alegre: Edipucrs; p. 487–491.
- Arcila D, Orti G, Vari R, Armbruster JW, Stiassny MLJ, Ko KD, Sabaj MH, Lundberg J, Revell LJ, Betancur-R R. 2017. Genome-wide interrogation advances resolution of recalcitrant groups in the tree of life. Nat Ecol Evol. 1:0056.
- Eschmeyer WN, Fricke R, van der Laan R. (eds). 2018. Catalog of fishes: Sternopygidae. [accessed 2018 Feb 27]. http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 17:754–755.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. Mitofish and mitoannotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
