Abstract
The acanthocephalan Hebesoma violentum Van Cleave was obtained in the intestine of Siniperca chuatsi. The complete mt genome sequence of H. violentum was obtained by long PCR, containing 36 genes with 12 protein coding genes, 22 transfer RNAs (tRNAs), and two ribosomal RNAs (rRNAs).
Keywords:
Acanthocephalans were recently recognized as a group within the phylum Rotifera in either traditional morphological (Sørensen and Giribet Citation2006) or molecular phylogenetic analyses (García-Varela and Nadler Citation2006). In this study, the complete mt genome of Hebesoma violentum was sequenced.
Hebesoma violentum was dissected out from intestines of Siniperca chuatsi, which were captured in Liangzi Lake (30°08′58.1″N, 114°67′36.3″E). The specimen is stored in Fisheries Institute, Anhui Academy of Agriculture Sciences. We amplified mitochondrial DNA using the long PCR method as shown in Pan and Nie (Citation2013). The complete mitochondrial genome of H. violentum is 13,393 bp (KC415004).
The genome of H. violentum is encoded on the same strand and in the same direction, and they all contain a total of 36 genes and 12 protein-coding genes, including 22 transfer RNAs and two ribosomal RNAs. The 12 protein-coding genes in the mt genome of H. violetum also share features in start and stop codons with those in other acanthocephalans.
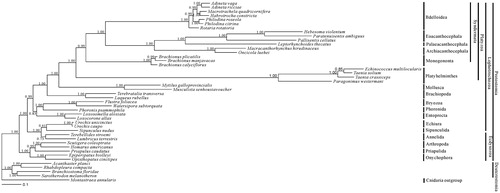
Phylogenetic analysis was performed using nine of the 12 protein-coding genes by Bayesian inference (BI). The clade containing acanthocephalans and bdelloids is well revealed with high supporting value in the phylogenetic tree. It is indicated that Eoacanthocephala and Palaeacanthocephala are closely related, forming a clade, with Archiacanthocephala as a sister group ().
Figure 1. Bayesian phylogenetic tree inferred from amino acid sequence dataset of nine protein-coding genes for 44 metazoan mitochondrial genomes. The tree shows the topology based on concatenated data of nine mitochondrial encoded protein sequences (cox1, atp6, nad4L, nad4, nad5, cob, nad1, cox2, cox3). Reconstruction was performed by MrBayes version 3.2. The numerical values near internal nodes represent Bayesian posterior probability (BPP) values.
Acknowledgements
We thank Dr Maocang Yan for his comments on the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- García-varela M, Nadler SA. 2006. Phylogenetic relationships among Syndermata inferred from nuclear and mitochondrial gene sequences. Mol Phylogenet Evol. 40:61–72.
- Pan TS, Nie P. 2013. The complete mitochondrial genome of Pallisentis celatus (Acanthocephala) with phylogenetic analysis of acanthocephalans and rotifers. Folia Parasitol. 60:181–191.
- Sørensen MV, Giribet G. 2006. A modern approach to rotiferan phylogeny: combining morphological and molecular data. Mol Phylogenet Evol. 40:585–608.
