Abstract
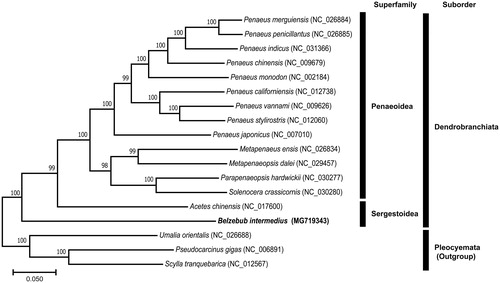
We determined the mitochondrial genome (mitogenome) sequence of Belzebub intermedius (Hansen, 1919). To the best of our knowledge, this is the first complete mitogenome in the family Luciferidae. The complete mitogenome of B. intermedius is 16,001 bp in length, with 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs), and a control region (CR). The gene arrangement of B. intermedius is almost identical to that of the same sergestoid species, Acetes chinensis Hansen, 1919, except that there is no additional trnS1. A maximum-likelihood tree, constructed using 18 decapod mitogenomes, confirmed that B. intermedius occupied the most basal position within the suborder Dendrobranchiata.
Dendrobranchiata, a suborder of the decapod shrimps (commonly known as prawns), comprises two superfamilies, Sergestoidea and Penaeoidea. The Sergestoidea is divided into two families, Luciferidae and Sergestidae. The Luciferidae was considered monotypic with only the genus Lucifer. However, Belzebub Vereshchaka, Olesen & Lunina, 2016 has been established as a new genus through morphological cladistic analysis. Therefore, Luciferidae is currently divided into two genera (Vereshchaka et al. Citation2016). The Belzebub consists of five species, of which B. intermedius is considered a euryhaline and a subtropical pelagic species with adaption to high temperatures (Xu Citation2010; WoRMS Citation2018). Furthermore, the distribution and abundance of B. intermedius are known to affect the weather, global warming, and the Kuroshio currents in both of the East China Sea and the Yellow Sea (Ma et al. Citation2009). In the present study, we determined the complete mitogenome of B. intermedius. To the best of our knowledge, this is the first complete mitogenome in Luciferidae. Therefore, it would be useful and essential for further studies on phylogenetic relationships and the evolution of Dendrobranchiata, including Sergestoidea.
A single specimen of B. intermedius was collected using a Norpac net from the Eulwang-ri beach (37°26′N, 126°22′E) in the Yellow Sea, South Korea. Specimen was deposited in the National Institute of Biological Resources, Incheon, South Korea (specimen deposit no. NIBRIV0000816436). Mitochondrial DNA extraction, sequencing and gene annotation were performed according to the methods described by Song et al. (Citation2016). Commercial software Geneious version 6.1.3 (Biomatters Ltd, Auckland, New Zealand) was used for de novo assembly of raw reads and producing a circular form of the complete mitogenome with average coverage. The gene annotation was carried out using the MITOS (Bernt et al. Citation2013) and ARWEN (Laslett and Canbäck Citation2008). A phylogenetic tree was constructed using the software MEGA7.0 by the maximum-likelihood method (Kumar et al. Citation2016).
The complete mitogenome of B. intermedius (GenBank accession no. MG719343) is a circular molecule of 16,001 bp in length, with 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs), and a control region (CR). The gene content and arrangement of B. intermedius are almost identical to those of the same sergestoid species A. chinensis, except that there is no additional trnS1 (see Kim et al. Citation2012 for information of additional trnS1).
To infer the phylogenetic relationships, we performed a maximum-likelihood analysis using the concatenated sequences of 13 PCGs from 18 complete mitogenomes of decapods, including two sergestoid and 13 penaeoid species. Furthermore, three species of the suborder Pleocyemata were used as outgroups. As a result, B. intermedius occupied the most basal position within Dendrobranchiata ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Kim S, Kim J, Choi H-G, Park J-K, Min G-S. 2012. Complete mitochondrial genome of the northern mauxia shrimp Acetes chinensis (Decapoda, Dendrobranchiata, Sergestoidae). Mitochondrial DNA. 23:28–30.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Ma Z, Xu Z, Zhou J. 2009. Effect of global warming on the distribution of Lucifer intermedius and L. hanseni (Decapoda) in the Changjiang estuary. Prog Nat Sci. 19:1389–1395.
- Song J-H, Kim S, Shin S, Min G-S. 2016. The complete mitochondrial genome of the mysid shrimp, Neomysis japonica (Crustacea, Malacostraca, Mysida). Mitochondrial DNA A. 27:2781–2782.
- Vereshchaka AL, Lunina AA, Olesen J. 2016. Phylogeny and classification of the shrimp genera Acetes, Peisos, and Sicyonella (Sergestidae: Crustacea: Decapoda). Zool J Linn Soc-Lond. 177:353–377.
- WoRMS. 2018. Belzebub Vereshchaka, Olesen & Lunina, 2016; [accessed 2018 February 12]. http://www.marinespecies.org/aphia.php?p=taxdetails&id =883864.
- Xu ZL. 2010. Determining optimal temperature and salinity of Lucifer (Dendrobranchiata: Sergestoidea: Luciferidae) based on field data from the East China Sea. Plank Benthos Res. 5:136–143.