Abstract
We report the complete mitogenome of the common dolphin, Delphinus delphis. Overall structure of the 16,387 bp mitogenome was very similar to those of other delphinid species, including the ancient D. delphis individuals. Multigene phylogeny revealed that D. delphis was most closely related to Stenella coeruleoalba, and clustered well with other species within the subfamily Delphininae.
Although the genus Delphinus (Linnaeus, 1758) known to contain two species, the long-beaked form (D. capensis) is currently synonymized with D. delphis (Perrin Citation2018). This species is widely distributed in the Atlantic and Pacific Oceans with two distinct subpopulations in the Mediterranean and Black Sea (Perrin Citation2002; Hammond et al. Citation2008). At present, D. delphis is listed in Appendix II of the Convention on International Trade in Endangered Species (CITES Citation2014) and has been listed as ‘Least Concern’ by the International Union for Conservation of Nature and Natural Resources Red List of Threatened Species (Hammond et al. Citation2008). A recent study revealed several mitogenomes of ancient D. delphis individuals excavated in the Black sea (Biard et al. Citation2017); however, the mitogenomes from existing individuals have not yet been reported. We report the complete mitogenome of an existing D. delphis individual.
Muscle tissue sample was collected from the frozen carcass of an adult male D. delphis (voucher no. CRI007520) deposited in the Cetacean Research Institute, National Institute of Fisheries Science that was found lifeless among the bycatch of gillnet fisheries in Mar 2017 along the South Sea (South Korea, 33°13′25″N 126°25′42″E). The D. delphis mitogenome was obtained by designating 15 primer pairs from the mitogenome of D. capensis (NC_012061.1) (Lee et al. Citation2018), and the sequences obtained using PCR amplifications were assembled and annotated as previously described (Kim et al. Citation2017).
The D. delphis mitogenome (MH000365) was 16,387 bp in length (61.3% A + T content), and included a typical set of 37 genes (two rRNAs, 22 tRNAs, and 13 PCGs), and overall gene arrangement and content of the mitogenome were almost identical to those from other related Delphinidae species. The mitogenomic relatedness between D. delphis and other Delphinidae species was respectively determined using the ANI calculator (http://www.ezbiocloud.net/tools/ani). Although the mitogenomes from two ancient D. delphis individuals (Biard et al. Citation2017) were the most similar to CRI007520 with 99.6% each, the highest species-specific OrthoANI value was obtained for Stenellacoeruleoalba (NC_012053, 98.3%), whereas the lowest was for killer whale (Orcinus orca; NC_023889, 93.0%).
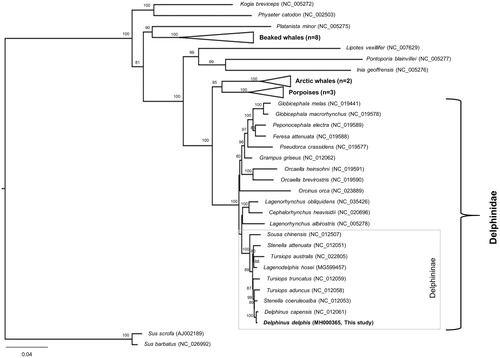
The taxonomical position of D. delphis was determined using multigene phylogenetic analysis (Kim et al. Citation2017). The resultant phylogeny suggested that D. delphis was most related to S. coeruleoalba and clustered well with other dolphin species within the subfamily Delphininae (Rosel et al. Citation1994; LeDuc et al. Citation1999; Amaral et al. Citation2007; McGowen Citation2011; Amaral et al. Citation2012) (). The first decoded complete mitogenome of D. delphis and its associated genomic information might provide important insights for its conservation and further evolutionary studies within the subfamily Delphininae.
Figure 1. Maximum-likelihood phylogeny based on the 13 concatenated PCGs from the available Odontoceti mitogenomes. The grey square box denotes the species currently placed in the subfamily Delphininae. Numbers at the branches indicate bootstrapping values obtained with 1000 replicates, and only bootstrap values >70% are indicated. The scale bar represents 0.04 nucleotide substitutions per site. D. capensis is currently synonymized with D. delphis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Amaral AR, Jackson JA, Möller LM, Beheregaray LB, Coelho MM. 2012. Species tree of a recent radiation: the subfamily Delphininae (Cetacea, Mammalia). Mol Phylogenet Evol. 64:243–253.
- Amaral AR, Sequeira M, Martínez-Cedeira J, Coelho MM. 2007. New insights on population genetic structure of Delphinus delphis from the northeast Atlantic and phylogenetic relationships within the genus inferred from two mitochondrial markers. Mar Biol. 151:1967–1976.
- Biard V, Gol'din P, Gladilina E, Vishnyakova K, McGrath K, Vieira FG, Wales N, Fontaine MC, Speller C, Olsen MT. 2017. Genomic and proteomic identification of Late Holocene remains: setting baselines for Black Sea odontocetes. J Archaeol Sci Rep. 15:262–271.
- Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES). 2014. Appendices I, II and III; [accessed 2018 Mar 27]. http://www.cites.org/eng/app/appendices.php
- Hammond PS, Bearzi G, Bjørge A, Forney K, Karczmarski L, Kasuya T, Perrin WF, Scott MD, Wang JY, Wells RS, et al. 2008. Delphinus delphis. The IUCN Red List of Threatened Species 2008:e.T6336A12649851; [accessed 2018 Mar 27]. http://dx.doi.org/10.2305/IUCN.UK.2008.RLTS.T6336A12649851.en
- Kim JH, Lee YR, Koh JR, Jun JW, Giri SS, Kim HJ, Chi C, Yun S, Kim SG, Kim SW, et al. 2017. Complete mitochondrial genome of the beluga whale Delphinapterus leucas (Cetacea: Monodontidae). Conserv Genet Resour. 9:435–438.
- LeDuc RG, Perrin WF, Dizon AE. 1999. Phylogenetic relationships among the delphinid cetaceans based on full cytochrome b sequences. Mar Mamm Sci. 15:619–648.
- Lee K, Lee J, Cho Y, Kim HW, Park KJ, Sohn H, Choi Y, Kim HK, Jeong DG, Kim JH. 2018. First report of the complete mitochondrial genome and phylogenetic analysis of Fraser’s dolphin Lagenodelphis hosei (Cetacea: Delphinidae). Conserv Genet Resour. doi:10.1007/s12686-017-0964-1.
- McGowen MR. 2011. Toward the resolution of an explosive radiation—a multilocus phylogeny of oceanic dolphins (Delphinidae). Mol Phylogenet Evol. 60:345–357.
- Perrin WF. 2002. Common dolphins Delphinus delphis, D. capensis, and D. tropicalis. In: Perrin WF, Würsig B, Thewissen JGM, editors. Encyclopedia of marine mammals. San Diego (CA): Academic Press; p. 245–248.
- Perrin WF. 2018. World Cetacea Database. Delphinus capensis Gray, 1828; [Accessed 2018 Mar 27]. http://www.marinespecies.org/aphia.php?p=taxdetails&id=137093
- Rosel PE, Dizon AE, Heyning JE. 1994. Genetic analysis of sympatric morphotypes of common dolphins (genus Delphinus). Mar Biol. 119:159–168.
