Abstract
Chloroplast (cp) genome sequences become a useful popular tool for population and phylogeny in recent reports. Here, the complete cp genome of the Euonymus fortunei has been reconstructed from the whole-genome Illumina sequencing data. The circular genome is 157,639 bp in size, and comprises a pair of inverted repeat (IR) regions of 26,668 bp each, a large single copy (LSC) region of 85,956 bp and a small single copy (SSC) region of 18,347 bp. The total GC content is 37.2%, while the corresponding values of the LSC, SSC, and IR region are 35.1%, 31.7%, and 42.7%, respectively. The cp genome contains 126 genes, including 89 protein-coding genes, eight ribosomal RNA genes, and 29 transfer RNA genes and one pseudogene. The maximum-likelihood phylogenetic analysis showed a strong sister relationship with Euonymus japonicus in Celastraceae. Our findings provide a foundation for further investigation of cp genome evolution in E. fortunei and other higher plants.
The chloroplast (cp) contains its autonomously replicating DNA genome that proceeds with photosynthesis and many other biological activities such as synthesizing starch, fatty acids, and other proteins relative to its special functions (Ohyama et al. Citation1986; Bausher et al. Citation2006). Most cp genomes are circular DNA molecules which have the typical quadripartite structure containing two repeat regions (IRa and IRb), LSC, and SSC (Yang et al. Citation2010).
Euonymus fortunei (Celastraceae), an evergreen shrub, is native to East Asia including China, Korea, and Japan. As an ornamental plant, E. fortunei has a wide range of applications in vertical garden and soil and water conservation. In addition, E. fortunei also serves as a traditional medicine for the treatment of inflammation and neurological diseases (Jian Citation2007; Zuo et al. Citation2012). To facilitate its genetic studies and thus contribute to its development and sustainable utilization, we assembled its cp genome using high-throughput Illumina sequencing technology in this study, as well as analysed its phylogenetic evolution, which will be helpful for better understanding of evolution within the Celastraceae and further studies on its molecular breeding and genetic engineering.
DNA extraction form the fresh leaves was collected from a single individual of E. fortunei in Xi’an Botanical Garden (Xi’an, Shaanxi Province, China). High-throughput DNA sequencing was conducted on the Illumina HiSeq 2500 Sequencing System (Illumina, San Diego, CA) by Breeding Biotechnologies (Breeding, Yangling, China). Total 16.8 M 150 bp raw reads were retrieved and trimmed by CLC Genomics Workbench v8.0 (CLC Bio, Aarhus, Denmark). A subset of 12.6 M trimmed reads were used for reconstructing the cp genome by NOVOPlasty (Dierckxsens et al. Citation2016), with that of its congener Euonymus schensianus (GenBank: NC_036019.1) as the initial reference genome. A total of 23,125,435 individual cp reads yielded an average coverage of 594.8-fold. The cp genome was annotated in GENEIOUS R9 (Biomatters Ltd., Auckland, New Zealand) by aligning with that of E. schensianus (NC_036019.1) and was drawn to the circular cp genome sequence map of OGDRAW 1.1.
The cp genome of E. fortunei is a double-stranded circular DNA molecule with 157,639 bp in size (MH150885). It comprises a pair of inverted repeat (IR) regions of 26,668 bp each, separated by a large single copy (LSC) region of 85,956 bp and a small single copy (SSC) region of 18,347 bp. The total GC content is 37.2%, while the corresponding values of the LSC, SSC, and IR region are 35.1%, 31.7%, and 42.7%, respectively.
This cp genome harbours 126 functional genes, including 89 protein-coding genes (PCGs), 29 tRNA genes, and eight rRNA genes. Eighteen PCGs, eight tRNA genes, and all rRNA genes are duplicated in the IR regions. The LSC region possesses 59 PCGs and 20 tRNA genes, while the SSC region contains 12 PCGs and one tRNA gene. Thirty three PCGs, 14 tRNA genes, and four rRNA genes are located in the forward strand while others are located in the reverse strand. Moreover, 14 genes contain one intron, while ycf3 harbours two introns; all the other genes are intronless. Among those genes, 44 are involved in photosynthesis, and 53 genes are involved in self replication. This is similar to those previously reported for the cp genomes of most other vascular plants (Yang et al. Citation2014).
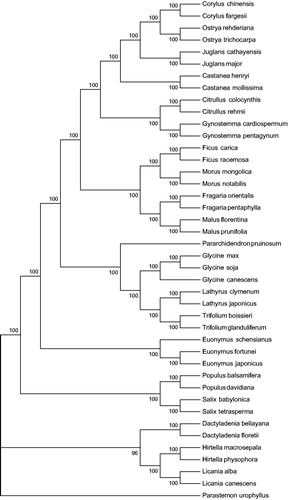
Sixty PCGs data among 41 cp sequences were aligned by MAFFT (Katoh et al. Citation2002) and then were connected as gene strings. The maximum-likelihood phylogenetic tree of E. fortunei was generated using those gene strings sequence by MEGA 6.0 (Tamura et al. Citation2013) with using 500 bootstrap replicates (). The phylogenetic analysis showed the position of E. fortunei was situated as the sister of E. japonicus and E. schensianus in Celastraceae. Our findings provide a foundation for further investigation of cp genome evolution in E. fortunei and other higher plants.
Figure 1. Phylogenetic data of 41 species within the family Celastraceae based on the maximum-likelihood analysis of the whole chloroplast genome sequences using 500 bootstrap replicates. The analysed species and corresponding GenBank accession numbers are as follows: Corylus chinensis NC_032351.1, Corylus fargesii NC_031854.1, Castanea henryi NC_033881.1, Castanea mollissima NC_014674.1, Citrullus colocynthis NC_035727.1, Citrullus rehmii NC_035975.1, Dactyladenia bellayana NC_030555.1, Dactyladenia floretii NC_030557.1, Euonymus japonicus NC_028067.1, Euonymus schensianus NC_036019.1, Fragaria orientalis NC_035501.1, Fragaria pentaphylla NC_034347.1, Ficus carica NC_035237.1, Ficus racemosa NC_028185.1, Glycine canescens NC_021647.1, Glycine max NC_007942.1, Glycine soja NC_022868.1, Gynostemma cardiospermum NC_035959.1, Gynostemma pentagynum NC_036136.1, Hirtella macrosepala NC_030561.1, Hirtella physophora NC_024066.1, Juglans cathayensis NC_033893.1, Juglans major NC_035966.1, Juglans cathayensis NC_033893.1, Juglans major NC_035966.1, Lathyrus clymenum NC_027148.1, Lathyrus japonicus NC_027075.1, Licania alba NC_024064.1, Licania canescens NC_030566.1, Malus florentina NC_035625.1, Malus prunifolia NC_031163.1, Morus mongolica NC_025772.2, Morus notabilis NC_027110.1, Ostrya rehderiana NC_028349.1, Ostrya trichocarpa NC_034295.1, Pararchidendron pruinosum NC_035348.1, Parastemon urophyllus NC_030517.1, Populus balsamifera NC_024735.1, Populus davidiana NC_032717.1, Salix babylonica NC_028350.1, Salix tetrasperma NC_035744.1, Trifolium boissieri NC_025743.1, and Trifolium glanduliferum NC_025744.1.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bausher MG, Singh ND, Lee SB, Jansen RK, Daniell H. 2006. The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var ’Ridge Pineapple‘: organization and phylogenetic relationships to other angiosperms. BMC Plant Biol. 6:21.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Jian X. 2007. Effect of Euonymus fortunei extract on the expression of c-fos in rats with cerebral ischemia-reperfusion injury. Guangxi Med J. 10:011.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, Umezono K, Shiki Y, Takeuchi M, Chang Z, et al. 1986. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 322:572.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031.
- Yang M, Zhang X, Liu G, Yin Y, Chen K, Yun Q, Zhao D, Al-Mssallem IS, Yu J. 2010. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.). PLoS One. 5:e12762.
- Zuo GY, Zhang XJ, Yang CX, Han J, Wang GC, Bian ZQ. 2012. Evaluation of traditional Chinese medicinal plants for anti-MRSA activity with reference to the treatment record of infectious diseases. Molecules. 17:2955–2967.
