Abstract
In this study, the complete mitochondrial genome of Epinephelus akaara × Epinephelus lanceolatus has been presented. The mitochondrial genome is 16,795 bp long and consists of 13 protein-coding genes, 2 rRNA, 22 tRNA, and a D-loop region. The phylogenetic analysis by neighbour-joining (MJ) method showed that the hybrid grouper has the closer relationship to E. akaara.
Epinephelus akaara and E. lanceolatus are both belong to Serranidae in the ordo perciformes. Both of them have high economic value and are widely distributed in South China, Korea, southern China, and Southeast Asian countries. The hybrid grouper F1 is inclined to take E. akaara as female parent and E. lanceolatus as male parent. As the hybrid generation has a faster growth and a better disease resistance, and there is little information of F1 genetic characteristics, the next-generation sequencing technique was used to analysis the complete mitochondrial genome. The specimen of F1 generation was obtained from the Marine Fisheries Development Center of Guangdong Province, China. The total genomic DNA was extracted from the fin of the fresh fish using the salting-out procedure (Howe et al. Citation1997).
The complete mitochondrial genome of E. akaara × E. lanceolatus is 16,795 bp in length (Genebank, KY132101), consisting of 13 protein-coding genes, 2 rRNA, 22 tRNA, and a D-loop region. Most of the genes are encoded on the heavy strand, with only the DADH dehydrogenase subunit 6 and eight tRNA genes are encoded on the light strand. The composition nucleotide of light strand is 27.28% for A, 28.68% for T, 27.84% for G, and 16.2% for C, and the heavy strand is 28.68% for A, 27.28% for T, 16.20 for G, and 27.84 for C.
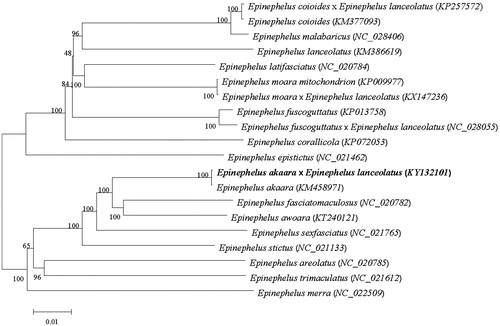
We perform multiple sequence alignment and construct a neighbour-joining (MJ) phylogenetic tree () (Tamura et al. Citation2013). As shown, the hybrid grouper was closer with the E. akaara.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Howe JR, Klimstra DS, Cordon-Cardo C. 1997. DNA extraction from paraffin-embedded tissues using a salting-out procedure: a reliable method for PCR amplification of archival material. Histol. Histopathol. 12:595–601.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.