Abstract
The first complete chloroplast genome sequences of Korean endemic basswood in Ulleung Island, Tilia insularis, were reported in this study. The T. insularis plastome was 162,565 bp long, with the large single-copy (LSC) region of 91,100 bp, the small single-copy (SSC) region of 20,381 bp, and two inverted repeat (IR) regions of 25,542 bp. The plastome contained 131 genes, including 86 protein-coding, eight ribosomal RNA, and 37 transfer RNA genes. The overall GC content was 36.5%. Phylogenetic analysis of 14 representative plastomes within the family Malvaceae suggests that T. insularis is closely related to other congeneric species in subfamily Tilioideae.
The basswood genus Tilia L. belongs to subfamily Tilioideae of Malvaceae and consists of 23 species widely distributed in the temperate northern hemisphere (Alverson et al. Citation1999; APG III Citation2009; Pigott Citation2012). Tilia is an ecologically important genus as one of the dominant deciduous tree species in the broad-leaved temperate forests and economically as useful timbers, honey resources, and ornamental trees (Pigott Citation2012; Cai et al. Citation2015; Bu and Park Citation2016). Of 23 species of Tilia, nine native and two cultivated species are known in Korea (Chung and Kim Citation1983; Kim and Chung Citation1986). Of nine native species, T. insularis Nakai is endemic to young volcanic Ulleung Island, which is located approximately 130 km east of the Korean peninsula (Nakai Citation1917; Kim Citation1985). Taxonomic status of T. insularis and its relationship to other species have long been controversial. For example, Yoon et al (Citation2003) investigated the vegetative and reproductive morphology among T. amurensis, T. insularis, and T. taquetii in Korea and T. japonica in Japan, and concluded that the morphological differentiation among them were not sufficient enough to be recognized as distinct species. A similar morphological study between T. amurensis and T. insularis by Pigott (2008) also concluded that the two should be treated as conspecific. However, recent molecular phylogenetic study suggested that T. insularis is more closely related to T. japonica than T. amurensis (Bu and Park Citation2016). It is still required to determine the taxonomic status of T. insularis and its phylogenetic position (Chung and Kim Citation1983; Choi Citation2008). In this study, we sequenced the complete chloroplast genome of T. insularis and compared it to congeneric species.
Total DNA (Voucher specimen: KNU-Lee171010) was isolated using the DNeasy plant Mini Kit (Quiagen, Carlsbad, CA) and sequenced by the Illumina HiSeq 4000 (Illumina Inc., San Diego, CA). A total of 31,656,366 paired-end reads were obtained and assembled de novo with Velvet v. 1.2.10 using multiple k-mers (Zerbino and Birney Citation2008). The tRNAs were confirmed using tRNAsacn-SE (Lowe and Eddy Citation1997). The total plastome length of T. insularis (MH169579) was 162,565 bp, with large single copy (LSC; 91,100 bp), small single copy (SSC; 20,381 bp), and two inverted repeats (IRa and IRb; 25,542 bp each). The overall GC content was 36.5% (LSC, 34.2%; SSC, 31.0%; IRs, 43.0%) and the plastome contained 131 genes, including 86 protein-coding, eight rRNA, and 37 tRNA genes. A total of 17 genes were duplicated in the inverted repeat regions including seven tRNA, four rRNA, and six protein-coding genes. The complete ycf1 gene was included in the IR at the SSC/IRa junction and the infA gene located in LSC became a pseudogene.
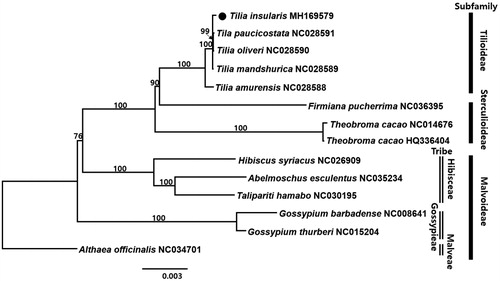
To confirm the phylogenetic position of T. insularis, 14 representative species of Malvaceae were aligned using MAFFT v.7 (Katoh and Standley Citation2013) and maximum likelihood (ML) analysis was conducted based on the concatenated 77 coding genes using IQ-TREE v.1.4.2 (Nguyen et al. Citation2015). The ML tree showed that T. insularis formed polytomy with T. manshurica, T. paucicostata, and T. oliveri.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alverson WS, Whitlock BA, Nyffeler R, Bayer C, Baum DA. 1999. Phylogenetic analysis of the core Malvales based on sequences of ndhF. Am J Bot. 86:1474–1486.
- APG III. 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 161:105–121.
- Bu D, Park SJ. 2016. Molecular phylogenetic study of Korean Tilia L. Korean J Plant Res. 29:547–554.
- Cai J, Ma P-F, Li H-T, Li D-Z. 2015. Complete plastid genome sequencing of four Tilia species (Malvaceae): a comparative analysis and phylogenetic implications. PLoS ONE. 10:e0142705
- Choi H-J. 2008. Morphology and phylogeny of Tilia [MSc Thesis]. Korea: Kangwon National University.
- Chung YH, Kim K-J. 1983. Monographic study of the endemic plants in Korea II. Taxonomy and interspecific relationships of the genus Tilia. Proc Coll Natur Sci SNU. 8:121–160. (in Korean).
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim KJ, Chung YH. 1986. A study of the distribution of genus Tilia. Korean J Apiculture. 1:24–25. (in Korean).
- Kim YK. 1985. Petrology of Ulreung volcanic island, Korea – part 1. Geology. J Japanese Assoc Mineral Petrol Econ Geol. 80:128–135.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Nakai T. 1917. Notulae et plantas Japoniae et Coreae, XIII. [Brief note of plant in Japan and Korea, XIII] Bot Mag Tokyo. 31:3–30.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Pigott CD. 2008. The identity of limes labelled Tilia insularis. The Plantsman (new series). 7:194–195.
- Pigott CD. 2012. Limes-tree and Basswoods a biological monograph of the genus Tilia. New York: Cambridge University Press.
- Yoon JG, Chang C-S, Kim H, Chang K-S, Lee H-S. 2003. Species delimitation of Tilia amuruensis, T. taquetii, and T, japonica. Bull Seoul Nat’l Univ Arboretum. 23:78–88. (in Korean).
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.