Abstract
Here, the complete mitogenome of one commercial Saccharina cultivar in Chinese market which marked ‘Shichang’ was reported. Circular-mapping mitogenome with an overall A + T content of 64.70% was 37,657 bp in size. It encoded three rRNAs (23s, 16s, and 5s), 25 tRNAs, 38 proteins (including three open reading frames, ORFs). Gene component and arrangement of ‘Shichang’ mitogenome were identical to those reported Saccharina cultivars. Further phylogenetic analysis of mitogenomes confirmed that ‘Shichang’ had closer relationship with cultivar ‘Pingbancai’, which suggested its parental origin and genetic relationship.
Saccharina (Laminariales, Phaeophyceae) is one of the most important seaweeds with its great economic value (Kain Citation1979). In China, about 20 cultivars have been bred (Zhang et al. Citation2016b), and most of them were selected from Saccharina japonica. However, parental origin of Saccharina cultivar in Chinese market is not clear due to the genetic hybrid during the breeding and cultivation. Here, we obtained the complete mitogenome of one commercial cultivar from the market of Weihai, Shandong, China (37°16′N, 122°41′E) and conducted phylogenetic analysis to provide new molecular data to evaluate the parental origin and genetic relationship of Chinese commercial Saccharina cultivar.
‘Shichang’ sample (specimen number: 201705002) was stored at –80 °C for DNA extraction. The protocol and data processing of this work were followed by Zhang et al. (Citation2011).
The complete mitogenome of ‘Shichang’ was characterized as a circular molecule of 37,657 bp (GenBank accession number MG712781). The nucleotide contained as follows: A = 10,700 (28.4%), T = 13,664 (36.29%), G = 7750 (20.58%), and C = 5543 (14.72%). The mitogenome had an overall A + T content of 64.70%, exhibiting a high AT richness. Cumulative AT-skew (–0.1218) and GC-skew (0.1660) analysis of mitogenome reflected a slight bias towards T and G in nucleotide composition on H-strand. The mitogenome encoded 66 genes including three rRNAs, 25 tRNAs, 35 protein-encoding genes, and three open reading frames (ORFs). Apart from rpl2, rpl16, rps3, rps19, tacC, and ORF130, 60 genes were encoded on H-strand. In this study, rps8–rpl6–rps2–rps4 was also found as a conserved gene cluster in those reported Saccharina species and cultivars. All protein-encoding genes used ATG as the initial codon and 68.42% used TAA as stop codon, higher than that for TAG (21.05%) and TGA (10.53%).
According to the total alignment of ‘Shichang’ and S. japonica, only 11 nucleotide mutations were detected and four led to the variety of amino acid. The result of this work indicated the component and arrangement of Saccharina species and cultivars mitogenomes had highly conservative evolution (Yotsukura et al. Citation2010; Zhang et al. Citation2011, Citation2013, Citation2016a, Citation2017).
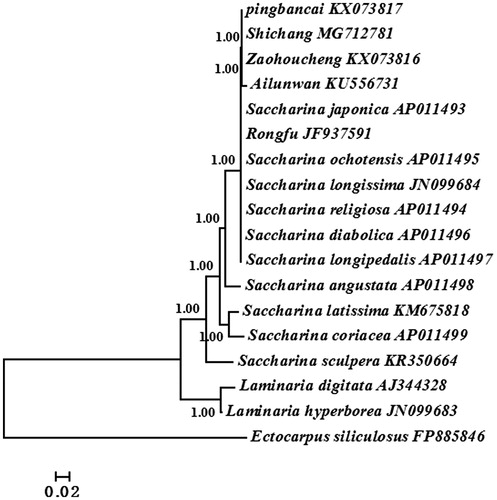
Bayesian method based on the whole mitogenome sequences of 17 available Saccharina and Laminaria algae was used to reconstruct the phylogenetic tree (). Ectocarpus siliculosus was selected as the outgroup. The results of phylogenetic analyses showed that all algae were divided into two groups: Saccharina and Laminaria. ‘Shichang’ together with other reported Saccharina cultivars were all in Saccharina clade and ‘Shichang’ had the closest relationship with cultivar ‘Pingbancai’. This study would facilitate us to better understand the parental origin and genetic relationship of commercial Saccharina cultivar in Chinese market.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kain JM. 1979. A view on the genus Laminarina. Oceanogr Mar Biol Annu Rev. 17:101–161.
- Yotsukura N, Shimizu T, Katayama T, Druehl LD. 2010. Mitochondrial DNA sequence variation of four Saccharina species (Laminariales, Phaeophyceae) growing in Japan. J Appl Phycol. 22:243–251.
- Zhang J, Gao BJ, Liu T, Liu N. 2016a. The complete mitogenome of “Pingbancai”, an important economic Saccharina cultivation variety. Mitochondrial DNA B: Resour. 1:470–471.
- Zhang J, Li N, Liu N, Liu T. 2017. The complete mitogenome of “Zaohoucheng”, an important Saccharina japonica cultivar in China. Mitochondrial DNA B: Resour. 2:656–657.
- Zhang J, Li N, Zhang ZF, Liu T. 2011. Structure analysis of the complete mitochondrial genome in cultivation variety “Rongfu”. J Ocean Univ China. 10:351–356.
- Zhang j, Liu T, Bian DP, Zhang L, Li XB, Liu DT, Liu C, Cui JJ, Xiao LY. 2016b. Breeding and genetic stability evaluation of the new Saccharina variety “Ailunwan” with high yield. J Appl Phycol. 28:3413–3421.
- Zhang J, Wang XM, Liu C, Jin YM, Liu T. 2013. The complete mitochondrial genomes of two brown algae (Laminariales, Phaeophyceae) and phylogenetic analysis within Laminaria. J Appl Phycol. 25:1247–1253.