Abstract
Citrus sinensis is an important agricultural product with huge economic value. In this study, we report the complete mitochondrial (mt) genome sequence of Citrus sinensis for the first time. Our assembly of the Citrus sinensis mt genome resulted in a final sequence of 640,906 bp in length which contains 63 genes. Phylogenetic analysis showed that Citrus sinensis was closely related to Brassica napus and Arabidopsis thaliana, which also belong to malvids. The complete mt genome will facilitate further genetic studies of Citrus sinensis.
Citrus sinensis, also known as sweet orange, is an important agricultural product of immense economic value. Citrus sinensis is a member of Rutaceae. The genus Citrus includes five major cultivated species, including Citrus sinensis (sweet orange), Citrus reticulata (tangerine and mandarin), Citrus limon (lemon), Citrus grandis (pummelo), and Citrus paradisi (grapefruit) (Xu et al. Citation2013). Moreover, oranges are significant nutritional source for human health.
The whole genomic DNA was extracted from leaves of a mature sweet orange tree which was growing in a citrus orchard in Dundee, Florida (28°01’N 81°37’W). Genome sequence was generated on 454 platforms. Fourteen different single-end shotgun libraries were prepared as well as several paired-end libraries with pair distances of 3 kb and 8 kb for sequencing on the 454 platform to ensure that coverage of different parts of the genome was as even as possible. A total of 51.5× of 454 sequencing data were generated (Wu et al. Citation2014). Mitochondrias are essential organelles in plants which produce the energy of the cell. In this study, we described the assembly and annotation details of the Citrus sinensis mt genome, which will provide help to the study of molecular identification, genetic diversity, and phylogenetic classification in Rutales. The complete mitochondrial (mt) genome was submitted to the GenBank under the accession number of NC_037463.
All the 454 GS FLX titanium raw reads were assembled using Newbler 2.7. The original contigs generated by Newbler were a mixture of nuclear and organellar DNA. For the purpose of isolating mitochondrion contigs from all the contigs we obtained, we used a method described by Wang et al. (Citation2018). According to Wang’s method, the coverage between 10 and 100 were preliminary set to filter the most likely mt contigs out. To visualize the connections among the filtered out mt contigs, we used Perl scripts and a text file named ‘454ContigGraph.txt’ which was generated by Newbler. Referring to the reads depth of the contigs and the connecting map, we removed false links and wrong forks manually. After this, a circular mt genome was developed.
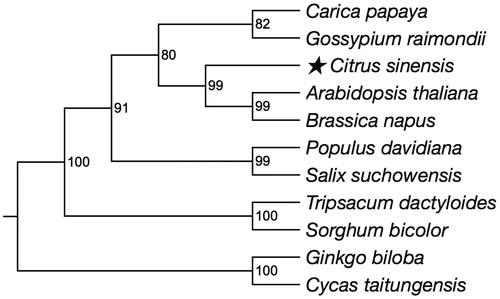
The complete Citrus sinensis mt genome is 640,906 bp in length. The overall G + C content of mt genome is 43.89%. The physical map of Citrus sinensis mt genome was generated with OGDRAW (Lohse et al. Citation2007). With the help of the online program MITOFY, a total of 63 genes were identified, including 32 protein-coding genes, 28 tRNAs, and three rRNAs. Most of these genes were single copy genes, while five genes existed as double copies, including one protein-coding gene (rpl2) and four tRNA genes (trnl-CAU, trnA-Val, trnA-Asp, trnA-Trp). Besides, tRNA gene trnA-Ser existed as quadra copies. Twenty two protein-coding genes were extracted from mt genomes of 16 species to construct the neighbour-joining phylogenetic tree (). A neighbour-joining phylogenomic analysis showed that Citrus sinensis was closely related to Brassica napus and Arabidopsis thaliana, which also belong to malvids.
Figure 1. The neighbour-joining phylogenetic tree of Citrus sinensis was constructed with MEGA 7 with 1000 bootstrap replicates using 22 protein-coding genes of 11 species. All the sequences used could be available in the GenBank database: Carica papaya NC_012116; Gossypium raimondii NC_029998; Citrus sinensis NC_037463; Arabidopsis thaliana NC_001284; Brassica napus NC_008285; Populus davidiana NC_035157; Salix suchowensis NC_029317; Tripsacum dactyloides NC_008362; Sorghum bicolor NC_008360; Ginkgo biloba NC_027976; Cycas taitungensis NC_010303.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52:267–274.
- Wang X, Cheng F, Rohlsen D, Bi C, Wang C, Xu Y, Wei S, Ye Q, Yin T. 2018. Organellar genome assembly methods and comparative analysis of horticultural plants. Hortic Res. 5:3.
- Wu GA, Prochnik S, Jenkins J, Salse J, Hellsten U, Murat F, Perrier X, Ruiz M, Scalabrin S, Terol J, et al. 2014. Sequencing of diverse mandarin, 1pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat Biotechnol. 32:656.
- Xu Q, Chen L-L, Ruan X, Chen D, Zhu A, Chen C, Bertrand D, Jiao WB, Hao BH, Lyon MP, et al. 2013. The draft genome of sweet orange (Citrus sinensis). Nat Genet. 45:59.
