Abstract
The complete chloroplast genome of Fargesia qinlingensis (Poaceae) has been reconstructed from the whole-genome Illumina sequencing data. The complete chloroplast genome sequence of F. qinlingensis obtained in this study was 139,640 bp in length, with a large single copy (LSC) region of 83,220 bp, a small single copy (SSC) region of 12,826 bp, separated by two inverted repeat (IR) regions of 21,797 bp each. The GC content was 38.9%, and in the LSC, SSC, and IR regions were 37.0%, 33.2%, and 44.2%, respectively. A total of 130 genes were annotated, including 83 protein-coding genes (PCGs), 39 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. The result of the phylogenetic analysis showed that F. qinlingensis was more closely related to the species of F. denudata.
Fargesia qinlingensis (Poaceae), a Chinese endemic species, is mainly found in a special ecological zone from the elevation of 1600–3000 m in Mt. Qinling, Shaanxi, China (Liang Citation2000). It is also acting as a crucial food for the Giant panda (Liu and Jin Citation2008), and the growth status and the productivity of F. qinlingensis have a significant effect on the present living situation of the Giant panda (Li et al. Citation2016). The complete chloroplast genome of F. qinlingensis (Poaceae) has been reconstructed using Illumina pair-end sequencing data. The specific goals of the present study were to present the complete chloroplast genome sequences of F. qinlingensis, and provide new data for studying phylogenetic relationship with other plants and conservation of the endemic species in Poaceae. The annotated chloroplast genome sequence of this species has been submitted to GenBank with the accession number MH117939.
Leaf materials from F. qinlingensis (Poaceae) were obtained from Mt. Qinling, Shaanxi, China (33°44′40.01″N, 107°49’15.85’’E). The voucher specimens of F. qinlingensis were deposited in Institute of Botany of Shaanxi Province. The complete chloroplast genome of F. qinlingensis was sequenced using Illumina HiSeq 2500 (Illumina, San Diego, CA), yielding 10,978,383 reads which were then quality trimmed with CLC Genomics Workbench 8 (CLC Bio, Aarhus, Denmark). The programs of MITObim v1.8 (GitHub, San Francisco, CA) (Hahn et al. Citation2013) and Geneious v 10.1.2 (Biomatters Ltd., Auckland, New Zealand) were respectively employed for assembled and annotation of chloroplast genome, and Fargesia spathacea (GenBank: JX513417) was used as the initial reference.
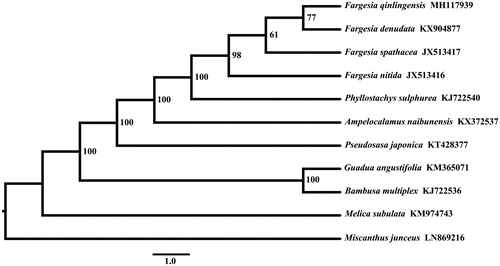
The chloroplast genome sequence of F. qinlingensis was 139,640 bp in length and composed of large single copy (LSC) region of 83,220 bp, small single copy (SSC) region of 12,826 bp and two inverted repeat (IR) copies 21,797 bp. The order and orientation of the gene arrangement pattern of F. qinlingensis was identical with that of F. spathacea. The GC content was 38.9%, and in the LSC, SSC, and IR regions were 37.0%, 33.2%, and 44.2%, respectively. A total of 130 genes were annotated, including 83 protein-coding genes (PCGs), 39 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. Eighteen genes (rps19, trnH-GUG, rpl2, rpl23, trnI-CAU, trnL-CAA, ndhB, rps7, trnV-GAC, rrn16, trnI-GAU, trnA-UGC, rrn23, rrn4.5, rrn5, trnR-ACG, trnN-GUU, rps15) were duplicated in the IR regions. For the trans-spliced rps12 gene, a 114bp 5′ end exon in the LSC region and a 3′ end with two exons (267 bp) were located in the IR region. Eight tRNA genes (trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU, trnA-UGU, trnA-UGC) and 10 PCGs (rps16, atpF, ydf3, rps12, petB, petD, rpl16, rpl2, ndhB, ndhA) contained one or two introns. The result showed that F. qinlingensis was more closely related to the species of F. denudata ().
Acknowledgements
We thank Lu Chao for the assistance in using RAxML.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Li YJ, Cai Q, Liu XH, Songer M, Wu PF, Jia XD, He XB. 2016. The effect of elevation on structure and nutrition of staple food bamboo and seasonal distribution of giant pandas. Acta Theriol Sin. 36:24–35.
- Liang TR. 2000. China forest. Beijing, China: Forestry Press; p. 1915–19l7.
- Liu XH, Jin XL. 2008. Habitat feature of giant pandas’ high-frequency activity areas on southern slope of the Qinling Mountains and habitat selection of giant panda. Chin J Ecol. 27:2123–2128.