Abstract
Prunus tomentosa belongs to subgenus Lithocerasus in family Rosaceae. It is native to northern China and has become a staple back yard garden plant in Russia and much of Eastern Europe. In this study, de novo assembly with low coverage whole genome sequencing facilitated to generate the complete chloroplast (cp) genome of P. tomentosa. The genome size is 158,356 bp in length. It exhibited a typical quadripartite structure comprising a large single copy region (LSC, 86,630 bp), a small single copy region (SSC, 19,010 bp) and a pair of inverted repeat regions (IRs, 26,358 bp each). A total of 115 genes were predicted including 82 protein-coding genes, 29 tRNA genes, and four rRNA genes. Phylogenetic analysis indicated that P. tomentosa is most closely related to P. mume suggesting the phylogenetic relationship between P. tomentosa and subgenus prunpphora.
Prunus tomentosa Thunb. (commonly Tomentosa cherry or Nanking Cherry) is an important deciduous fruit tree endowed with highly ornamental and economic values (Yü et al. Citation1986). It is native to northern China and naturalized in Japan, Russia, and other northern regions of the continent (Hamilton et al. Citation2016). For centuries, P. tomentosa has possessed many valuable traits, such as various peel colour, very cold tolerant, diverse environments adaption (Zhang et al. Citation2008). It can be used as a gene donor to improve other Prunus species. However, such breeding programs require a sufficient understanding of the groups’ phylogeny among different species (Zhang and Gu Citation2016). The phylogenetic relationship of P. tomentosa was always controverted (Shi et al. Citation2013). In this study, we generated the complete chloroplast (cp) genome sequence of P. tomentosa, which could help us verify the phylogenetic relationship between P. tomentosa and its relative species.
The plant material was obtained from Changyi, Shandong province, China (35°45.5146′ N, 119°26.4163′ E, altitude 25 m). Total DNA was extracted with a modified CTAB protocol. An Illumina paired-end (PE) library was constructed and sequenced using an Illumina HiSeq 2500 platform (Illumina, San Diego, CA). After quality trimming, a total of 2.35 Gb clean PE reads (Phred scores >20) were assembled using SOAPdenovo (Li et al. Citation2009). Contigs were ordered and merged with the cp sequence of Prunus persica (NC_04697). Further validation was also performed using manual correction. The plastom was annotated by Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. Citation2004) (http://dogma.ccbb.utexas.edu/).
Similarly, complete cp genome sequence of other 11 Rosales species (Pyrus pyrifolia NC015996 and Pyrus spinosa NC023130 as outgroups) were aligned using MAFFT 5 (Katoh et al. Citation2005). Maximum likelihood (ML) analysis was implemented in RAxML v8.2.4 (Stamatakis Citation2006). Maximum parsimony (MP) and neighbour-joining (NJ) analysis were performed using MEGA 6.0 (Tamura et al. Citation2013) (http://www.megasoftware.net/).
The circular genome of P. tomentosa is 158,356 bp in size, with overall GC content 36.85%. It comprises a large single copy (LSC) region of 86,630 bp, a small single copy (SSC) region of 19,010 bp and a pair of inverted repeat (IR) of 26,358 bp. A total of 115 unique coding regions were predicted, comprising 82 protein-coding genes, 29 tRNA genes, and four rRNA genes. Among all unique genes, 19 genes contain one intron, two genes (ycf3 and clpP) with two introns, and gene atpB has three introns. All the coding regions accounted for 58.35% of the whole genome. The genome sequence was deposited in GenBank with the accession number MF_624726.
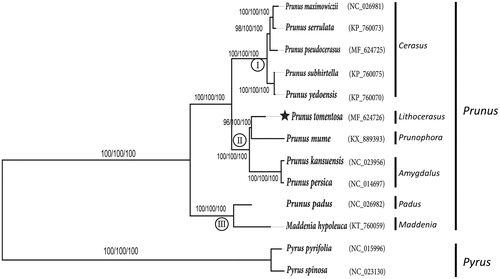
Phylogenetic analysis revealed three subgroups in genus Prunus (). Five species from subgenus Creasus formed one group, subgenus Padus and Maddenia composed of another clade. Interesting, P. tomentosa was nested within subgenus Amygdalus and prunpphora constituted the third group, which was, in turn, a sister to P. mume. This result was congruent with previous studies by isozyme (Shi et al. Citation2013) and other molecular markers (Chin et al. Citation2010).
Figure 1. Phylogenetic tree of P. tomentosa and other 12 species belonging to the Rosaceae. Tree was inferred from the complete chloroplast genome sequences using the ML method with a GTR model, MP method, and NJ method with a K-2P model. Only the framework of the ML tree was presented. Numbers in the nodes were the bootstrap values from 1000 replicates with an arrangement of ML/MP/NJ methods. Symbol (I, II, III) in the nodes represents three groups in genus Prunus.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chin SW, Wen J, Johnson G, Potter D. 2010. Merging Maddenia with the morphologically diverse Prunus (Rosaceae). Bot J Linn Soc. 164:236–245.
- Hamilton E, Maughan T, Black B. 2016. Nanking cherry in the garden. America: Utah State University Extension.
- Katoh K, Kuma KI, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucl Acids Res. 33:511–518.
- Li R, Yu C, Li Y, Lam T-W, Yiu SM, Kristiansen K, Wang J. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 25:1966–1967.
- Shi W, Wen J, Lutz S. 2013. Pollen morphology of the Maddenia clade of Prunus and its taxonomic and phylogenetic implications. J Syst Evol. 51:164–183.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yü T, Lu L, Ku T, Li C, Chen S. 1986. Rosaceae (3), Amygdaloideae. Flora Reipublicae Popularis Sinicae, vol. 38. Beijing (China): Science Press.
- Zhang Q, Gu D. 2016. Genetic relationships among 10 Prunus rootstock species from China, based on simple sequence repeat markers. J Am Soc Hortic Sci. 141:520–526.
- Zhang Q, Yan G, Dai H, Zhang X, Li C, Zhang Z. 2008. Characterization of tomentosa cherry (Prunus tomentosa Thunb.) genotypes using SSR markers and morphological traits. Sci Hortic. 118:39–47.
