Abstract
The whole cp genome of T. ciliata was 159,502 bp in length, containing a pair of inverted repeat (26,961 bp for each), a large single copy (87,199 bp) and a small single copy (18,381 bp) regions. The cp genome encoded 138 genes, including 89 protein-coding genes, 40 tRNA genes, 8 rRNA genes and 1 pseudogene. The nucleotide composition was asymmetric (30.7% A, 19.3% C, 18.6% G and 31.4% T) with an overall GC content of 37.9%. The maximum likelihood phylogenetic analysis based on 21 complete cp genome sequences showed that T. ciliata closely related to Azadirachta indica.
Toona ciliate is endemic to tropical and subtropical China. High value and huge potential use of T. ciliate directly incurred human overcutting and consequent habitat destruction during the last century, and thus, International Union for Conservation of Nature (IUCN) has included it in the Red List of Endangered species (Version 2.3, 2017). Some measurements have been taken to conserve this important species since 1991 in China (Zhao et al. Citation2015; Zhou et al. Citation2015). However, much still remained unknown about the complete chloroplast (cp) genome. In this study, we assembled the complete cp genome of T. ciliata based on the whole-genome Illumina sequencing dataset to improve an appreciation of its genetics that would be conducive to the formulation of conservation and management strategies.
Young leaves were collected from a single individual of T. ciliata at the campus of Fujian Agriculture and Forestry University (Fujian, China; 26°03′N, 119°16′E), and were dried using silica gel. Then, total genomic DNA was extracted using modified CTAB protocol (Yang et al. Citation2014) for the construction of a shotgun library, which was sequenced using the Illumina Hiseq 2000 Platform (Illumina, San Diego, CA). In total, 27,617,866 of 150 bp raw paired-end reads were obtained and the cp genome was assembled by MITObim v1.8 (Hahn et al. Citation2013) with the sequences of published Azadirachta indica (KF986530) cp genomes as the initial reference.
The cp genome of T. ciliata was annotated using software Geneious v 9.0.2 (Biomatters Ltd., Auckland, New Zealand) via comparison with the cp genome of relatively related species, including A. indica (KF986530), Leitneria floridiana (KT692940), and Litchi chinensis (KY635881). Finally, the physical map of the circular cp genome was generated using OGDRAW (Lohse et al. Citation2013).
The whole cp genome was 159,502 bp in length, containing a pair of inverted repeat (26,961 bp for each), a large single copy (87,199 bp) and a small single copy (18,381 bp) regions. The cp genome encoded 138 genes, including 89 protein-coding genes (77 unique), 40 tRNA genes (29 unique), eight rRNA genes (four unique), and one pseudogene. The nucleotide composition was asymmetric (30.7% A, 19.3% C, 18.6% G, and 31.4% T) with an overall GC content of 37.9%.
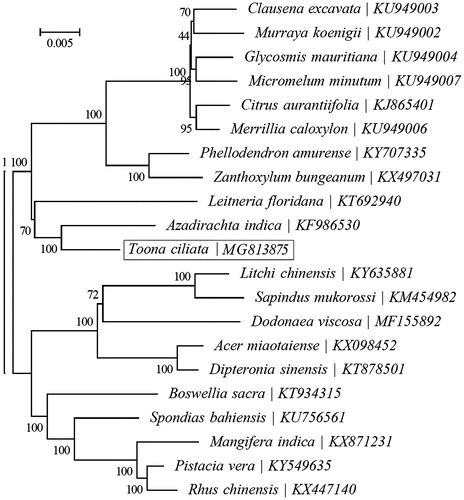
To investigate phylogenetic status of T. ciliate, a maximum likelihood (ML) analysis was reconstructed from the complete cp genomes of 19 species by the RAxML software (Stamatakis Citation2014) performed with 1000 replicates (). The phylogenetic analysis indicated that T. ciliata was closely related to A. indica. The complete cp genome of T. ciliata will supply useful genetic information for population genomic studies, and conservation management of this endangered species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031.
- Zhao BJ, Zhen AG, He RX, Cheng DH, Wang Y. 2015. Research on genetic physiological ecology of precious species: Toona ciliata and Toona ciliate var. pubescens. Agric Sci Technol. 16:997–1001.
- Zhou HM, Chen S, Fu CL, Xie CB, Li YJ, Wang Y. 2015. Research progress in cultivation and pharmaceutical chemicals of Toona ciliata var. pubescens and Toona ciliata. Agric Sci Technol. 16:722–726 + 760.