Abstract
Longchuanacris curvifurculus (L. curvifurculus) was once a dominating grasshopper in the Yunnan province (People’s Republic of China) that occupy important ecological niche. However, its population has severely declined because of the deterioration of ecological environment. Identifying the species and source of L. curvifurculus is important for biodiversity conservation and ecological/environmental preservation. In the study, the complete mitochondrial genome of L. curvifurculus was assembled from high-coverage (36.8×) Illumina MiSeq sequencing data. The circular genome is 15,450 bp in length, harboring 37 typical mitochondrial genes and one control region. The nucleotide composition is asymmetric (43.0% A, 14.3% C, 10.5% G, and 32.2% T), with an overall A + T content of 75.2%. All the protein-coding genes (PCGs) are initiated with typical ATN start codons and terminated by the typical TAA codons or the incomplete T(aa) codon. The control region has a remarkably high A + T content (84.9%) and is located between genes rrnS and trnV.
Longchuanacris curvifurculus (L. curvifurculus) is a type of short-horned grasshopper belonging to the order Orthoptera and family Catantopidae (Ramme Citation2008). Systematic studies of L. curvifurculus that lead to more effective and precise identification of this species, and will be important for biodiversity conservation and monitoring as well as for the management and control of population levels. Mitochondrial DNA, a powerful molecular tool for genetic research, has proven to be useful for such purposes (Song Citation2010; Zhao et al. Citation2010). In this investigation, the complete mitochondrial genome of L. curvifurculus has been assembled from whole-genome sequencing data generated using the Illumina MiSeq sequencing system (Illumina, San Diego, CA). The annotated mitogenomic sequence has been deposited into GenBank under the accession number MF989227.
An individual of L. curvifurculus was used in this study and the type sample of this species is now persevered in Museum of Da’li University, Yunnan, China. In all, 24.141 million raw reads were retrieved, quality-trimmed with Trimmomatic v0.35 (Bolger et al. Citation2014), and then used for the assembly of the mitochondrial genome with the Assembly by Reduced Complexity (ARC) pipeline (http://ibest.github.io/ARC/) (Hunter et al. Citation2015). This pipeline implements a hybrid mapping and assembly approach for targeted assembly of homologous sequences. A mitogenomic fragment (GenBank: EF437157) was used as the initial reference. About 3850 individual mitochondrial reads generated an average coverage of 36.8. Genome annotation was conducted using the MITOS Web Server (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013; Jansen et al. Citation2005; Papadakos et al. Citation2008), with precise adjustment via alignment with other available Catantopidae mitochondrial genomes (Liu and Qiu Citation2016; Yang et al. Citation2016).
The mitochondrial genome of L. curvifurculus is 15,450 bp in size, with a highly asymmetric nucleotide composition (43.0% A, 14.3% C, 10.5% G, and 32.2% T) and an overall A + T content of 75.2%. It harbours 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes (rrnS and rrnL), and one control region (). All 13 PCGs are initiated at typical ATN start codons, and are terminated with a TAA codon except nad5 and nad2 with the incomplete T(aa) codons. Sizes of the 22 tRNAs vary over a very small range, from 63 bp (trnC) to 71 bp (trnK and trnV). The two adjacent rRNAs are 1276 bp (rrnL) and 781 bp (rrnS) in length, with A + T contents of 77.60% and 74.20%, respectively. The control region, which is located between rrnS and trnV, is 617 bp long and has a remark-ably high A + T content (84.90%).The arrangement of genes is identical to that of other Catantopidae mitochondrial genomes (Hu et al. Citation2017).
Figure 1. Phylogenetic analysis using the 13 mitochondrial protein-coding genes of Longchuanacris curvifurculus. Mitochondrial genomes with the genbank accession ID of NC_033905, NC_033906, NC_032716, KX296781, NC_030586, NC_031397, NC_032303, KU355786, NC_026716, NC_031379, NC_031817, NC_013701, NC_027187, NC_023920, NC_013835, NC_013826, NC_036063, NC_021609, along with our data MF989227, were used to build the phylogenetic tree.
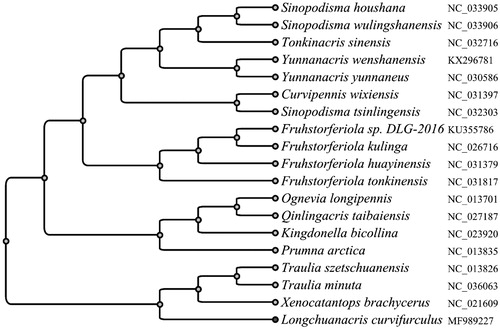
Using the sequences of 13 PCGs from the obtained mitochondrial genome of L. curvifurculus and 19 other Catantopidae/Melanoplinae mitochondrial genomes, an UPGMA tree was built to reveal the phylogenetic relationship among these species (). The results have shown that L. curvifurculus is close to genus Traulia and Xenocatantops, which are in consist with the previous morphology classifications.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Hunter SS, Lyon RT, Sarver BAJ, Hardwick K, Forney LJ, Settles ML. 2015. Assembly by Reduced Complexity (ARC): a hybrid approach for targeted assembly of homologous sequences. Biorxiv. 208:1-34.
- Hu Z, Guan DL, Mao BY. 2017. Description of a new species, caryanda stal, 1878 (acrididae, orthoptera) from china. Oriental Insects. 51:124–134.
- Jansen RK, Raubeson LA, Boore JL, dePamphilis CW, Chumley TW, Haberle RC, Wyman SK, Alverson AJ, Peery R, Herman SJ, et al. 2005. Methods for obtaining and analyzing whole chloroplast genome sequences. Meth. Enzymol. 395:348–384.
- Liu F, Qiu ZY. 2016. The complete mitochondrial genome of Fruhstorferiola huayinensis (Orthoptera: Catantopidae). Mitochondrial DNA Part B. 1:273–274.
- Papadakos P, Vasiliadis G, Theoharis Y, Armenatzoglou N, Kopidaki S, Marketakis Y, Daskalakis M, Karamaroudis K, Linardakis G, Makrydakis G. 2008. The anatomy of mitos web search engine. arXiv.
- Ramme W. 2008. Beiträge zur palaearktischen Orthopterenfauna (Tettigon. et Acrid.) II. Mitt Mus Nat kd Berl Zool Reihe. 18:416–434.
- Song H. 2010. Grasshopper systematics: past, present and future. J Orthoptera Res. 19:57–68.
- Yang J, Liu Y, Liu N. 2016. The complete mitochondrial genome of the Xenocatantops brachycerus (Orthoptera: Catantopidae). Mitochondrial DNA Part A. 27:2844–2845.
- Zhao L, Zheng Z, Huang Y, Sun H. 2010. A comparative analysis of mitochondrial genomes in Orthoptera (Arthropoda: Insecta) and genome descriptions of three grasshopper species. Zool Sci. 27:662–672.
