Abstract
In this study, we sequenced the complete mitochondrial genome of Pampus cinereus (Bloch, 1795). This mitochondrial genome, consisting of 16,540 base pairs (bp), contains 13 protein-coding genes, two ribosomal RNAs, 22 transfer RNAs, and two mainly noncoding regions (control region and origin of light-strand replication) as those found in other vertebrates. Control region with 846 bp in length, is located between tRNAPro and tRNAPhe. The overall base composition of the heavy strand shows 27.4% of T, 27.5% of C, 30.0% of A, and 15.2% of G, with a slight A + T rich bias (57.4%). The complete mitochondrial genome data will provide useful genetic markers for the studies on the molecular identification, population genetics, phylogenetic analysis and conservation genetics.
The grey pomfret, Pampus cinereus (Bloch, 1795) is a common species distributed mainly in the Indo-Western Pacific. This species is characterized by a greatly extended anal fin and notably long pectoral fins. However, some previous reports considered P. cinereus as a synonym of P. argenteus (Haedrich Citation1967; Parin and Piotrovsky Citation2004; Cui et al. Citation2011). To clarify the misidentification of this species, sufficient genetic information is needed and helpful for rational utilization of P. cinereus resources. In this study, we present the complete mitochondrial genome of P. cinereus and reconstruct phylogenetic topology of Pampus species to demonstrate the taxonomic status of P. cinereus.
The sample of P. cinereus was collected from the coastal waters of Xiamen (24.40°N, 118.16°E) during May 2012 and deposited in Fishery Ecology Laboratory of Ocean University of China (OUC). The complete P. cinereus mitogenome was amplified using a long-PCR technique, and then subsequent sequencing was accomplished by primer walking method (Miya and Nishida Citation1999). The mitogenome sequence has been deposited in GenBank with accession number MH037008.
The complete mitochondrial genome of P. cinereus (16,540 bp in length) consists of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and two mainly non-coding control region (control region and origin of light-strand replication). Arrangement of all genes is identical to that of most vertebrates (Wang et al. Citation2008; Chen Citation2013; Chiang et al. Citation2013). Most of the genes are encoded on the heavy strand (H-strand), except for the eight tRNA genes (-Gln, -Ala, -Asn, -Cys,-Tyr, -Ser, -Glu, and -Pro), one non-coding region (origin of light-strand replication, OL) and one protein-coding gene (ND6). The overall base composition is 27.4% for T, 27.5% for C, 30.0% for A, and 15.2% for G, with a slight A + T-rich feature (57.4%) just as other marine fishes (Cui et al. Citation2009; Cheng et al. Citation2012). Except for COI, ND4, and ND6 starting with GTG, the remaining 10 protein-coding genes start with ATG. Five protein-coding genes are inferred to terminate with incomplete stop codons (ND2, COII, COIII, ND3, ND4), with five (ATPase8, ATPase6, ND4L, ND5, and Cyt b) sharing TAA and three (ND1, COI, ND6) using TAG as stop codons. These features are common among vertebrate mitochondrial genome, and TAA is supposed to be appeared via posttranscriptional polyadenylation (Anderson et al. Citation1981).
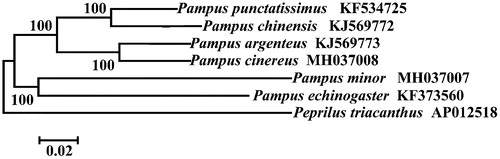
Phylogenetic relationship was constructed using NJ algorithm among six Pampus species based on 12 H-strand mitochondrial protein-coding genes, 22 tRNA and two rRNA genes (). These sequences were aligned using the DNASTAR software. MEGA 5.0 (Tamura et al. Citation2011) was then used to construct neighbour-joining (NJ) tree based on Kimura 2-parameter (K2P) model. Peprilus triacanthus (Perciformes: Stromateidae) was used as outgroup. Numbers above branches indicate neighbour-joining bootstrap percentages. Only bootstrap values of >50% are shown in the above NJ tree. This phylogenetic tree shows that these six Pampus species were obviously different from each other and P. cinereus is more closely related to P. argenteus than other species of Pampus species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290:457–465.
- Chen IS. 2013. The complete mitochondrial genome of Chinese sucker Myxocyprinus asiaticus (Cypriniformes, Catostomidae). Mitochondrial DNA. 24:680–682.
- Cheng J, Ma GQ, Song N, Gao TX. 2012. Complete mitochondrial genome sequence of bighead croaker Collichthys niveatus (Perciformes, Sciaenidae): a mitogenomic perspective on the phylogenetic relationships of Pseudosciaeniae. Gene. 491:210–223.
- Chiang TY, Chen IS, Lin HD, Chang WB, Ju YM. 2013. Complete mitochondrial genome of Sicyopterus japonicus (Perciformes, Gobiidae). Mitochondrial DNA. 24:191–193.
- Cui ZX, Liu Y, Li CP, Chu KH. 2011. Species delineation in Pampus (Perciformes) and the phylogenetic status of the Stromateoidei based on mitogenomics. Mol Biol Rep. 38:1103–1114.
- Cui ZX, Liu Y, Li CP, You F, Chu KH. 2009. The complete mitochondrial genome of the large yellow croaker, Larimichthys cracea (Perciformes, Sciaenidae): unusual features of its control region and the phylogenetic position of the Sciaenidae. Gene. 432:33–43.
- Haedrich RL. 1967. The Stromateoid fishes: systematics and a classification. Bull Museum Comp Zool. 135:31–139.
- Miya M, Nishida M. 1999. Organization of the mitochondrial genome of a deep-sea fish, Gonostoma gracile (Teleostei: Stomiiformes): first example of transfer RNA gene rearrangements in bony fishes. Mar Biotechnol. 1:416–426.
- Parin NV, Piotrovsky AS. 2004. Stromateoid fishes (suborder Stromateoidei) of the Indian Ocean (species composition, distribution, biology, and fisheries). J Appl Ichthyol. 44: 33–62.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Wang C, Chen Q, Lu G, Xu J, Yang Q, Li S. 2008. Complete mitochondrial genome of the grass carp (Teleostei, Cyprinidae, Gobioninae). Gene. 424:96–101.