Abstract
The complete circular mitochondrial genome of a higher termite Pericapritermes nitobei has a length of 15,224bp and encodes 37 genes including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA), 2 ribosomal RNA (rRNA) and a non-coding control region (D-loop). Protein coding genes (PCGs) in this circular mitogenome start with standard ATN initiation codons and end with complete termination codons TAN except for cox2 and nad5 genes with an incomplete stop codon T. The percentage of A and T (67.49%) is higher than that of G and C (32.51%). The phylogenetic tree revealed that mitogenomes of Pericapritermes formed one clade. The tree also revealed that Pericapritermes dolichocephalus and Pericapritermes latignathus constituted a sister group to P. nitobei. The date here provide a resource for genetics and evolution analysis within termites especially Pericapritermes genus.
Pericapritermes genus contains 33 species and mainly distributes in tropical and subtropical regions (Tan and Yan, Citation2013). As typical species of higher termites, the mitochondrial genomes of Pericapritermes are applied for termites evolutionary analysis (Bourguignon et al. Citation2015, Citation2017). Pericapritermes nitobei was first described as Eutermes nitobei and Krishna revised the species to P. nitobei (Krishna et al. Citation1968). Although there are some studies on the mitochondrial genes of P. nitobei such as rrnL, rrnS and cox2 (Ohkuma et al. Citation1999, Citation2004; Bujang et al. Citation2014), there is no available information about its complete mitochondrial genome. The present study was the first report on the complete mitochondrial genome sequences of P. nitobei.
Specimens were collected from Ningbo city, China and kept in the insect lab at Zhejiang A & F University with Accession number NB0024-TT-4. The entire mitochondrial genome sequence of P. nitobei was 15,224 bp in length, including 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes (rrnL and rrnS) and 1 non-coding control region (D-loop). Most of these genes were located on the H-strand except for nad1, nad4, nad4l, nad5 and 8 tRNA genes (trnQ, trnC, trnY, trnF, trnH, trnP, trnL1 and trnV). The genetic compositions and coding sequences are similar to other termites (Zhao et al. Citation2016; Hervé and Brune Citation2017).
The overall base compositions of P. nitobei mitochondrial genome had a high A/T tendency. The A + T content of P. nitobei was 67.49%, higher than that of G + C (32.51%). The mitochondrial genome of P. nitobei included intergenic spacers and overlapping regions. The intergenic spacer sequences were on 22 regions ranging in size from 1 to 17 bp. The overlapping sequences varied from 1 to 44 bp in five areas.
The mitochondrial genome of the P. nitobei contained 13 protein-encoding genes, with the total length of 11,144 bp. All protein-coding genes (PCGs) start with standard ATN initiation codons and end with complete termination codons TAN except for cox2 and nad5 genes with an incomplete stop codon T. There were 22 tRNA genes in the mitochondrial genome of P. nitobei. Except for the tRNA-Ser, which lacked the dihydrorubamide (DHU) arm, the other tRNAs have the classical cloverleaf structures.
The control region (D-loop) with 336 bp in size is located between rrnS and trnI gene. The content of A + T in control region was 70.54%, and its content was significantly higher than that of the whole mitochondrial genome.
Pericapritermes nitobei mitochondrial genome has two ribosomal RNAs, rrnL and rrnS. rrnL is located between trnL1 and trnV with the length of 1368 bp and rrnS is located between trnV and the control region with 806 bp in size.
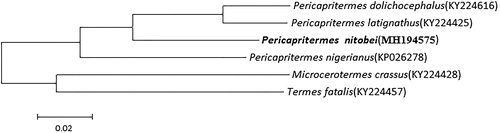
A phylogenetic tree among Pericapritermes was constructed using a dataset which contained all the nucleotide sequences of PCGs of Pericapritermes mitochondrial genes. Microcerotermes crassus and Termes fatalis (Termitidae) were set as outgroups. Two major clades were formed, one was M. crassus and T. fatalis, the other was the remaining species (Pericapritermes) (). The tree also revealed that P. dolichocephalus was closest to P. latignathus and constituted a sister group to P. nitobei.
Nucleotide sequence accession number
The complete genome sequence of P. nitobei has been assigned GenBank accession number (MH194575).
Acknowledgements
The authors thank for the financial support from NBTC-YYTC-ZAFU cooperative project (2045200283) and Developmental Fund of Zhejiang A&F University (2012FR087)
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Bourguignon T, Lo N, Cameron SL, Šobotník J, Hayashi Y, Shigenobu S, Watanabe D, Roisin Y, Miura T, Evans TA, et al. 2015. The evolutionary history of termites as inferred from 66 mitochondrial genomes. Mol Biol Evol. 32:406–421.
- Bourguignon T, Lo N, Šobotník J, Ho SYW, Iqbal N, Coissac E, Lee M, Jendryka MM, Sillam-Dussès D, Krížková B, et al. 2017. Mitochondrial phylogenomics resolves the global spread of higher termites, ecosystem engineers of the tropics. Mol Biol Evol. 34:589–597.
- Bujang NS, Harrison NA, Su NY. 2014. A phylogenetic study of endo-beta-1,4-glucanase in higher termites. Insectes Sociaux. 61:29–40.
- Hervé V, Brune A. 2017. The complete mitochondrial genomes of the higher termites Labiotermes labralis and Embiratermes neotenicus (Termitidae: Syntermitinae). Mitochondrial DNA Part B: Resources. 2:109–110.
- Krishna K, Grimaldi DA, Krishna V, Hayden T, Alfred E. 1968. Phylogeny and generic reclassification of the Capritermes complex (Isoptera, Termitidae, Termitinae). New York: Bulletin of the American Museum of Natural History; p. 293–294.
- Ohkuma M, Noda S, Kudo T. 1999. Phylogenetic relationships of symbiotic methanogens in diverse termites. Fems Microbiol Lett. 171:147–153.
- Ohkuma M, Yuzawa H, Amornsak W, Sornnuwat Y, Takematsu Y, Yamada A, Vongkaluang C, Sarnthoy O, Kirtibutr N, Noparatnaraporn N, et al. 2004. Molecular phylogeny of Asian termites (Isoptera) of the families termitidae and rhinotermitidae based on mitochondrial COII sequences. Mol Phylogenet Evol. 31:701–710.
- Tan SJ, Yan SH. 2013. A new species of the genus Pericapritermes silvestri (Isoptera, Termitidae) from Sichuan, China. Acta Zootaxon Sin. 38:119–123.
- Zhao S, Dang Y-L, Zhang H-G, Guo X-H, Su X-H, Xing L-X. 2016. The complete mitochondrial genome of the subterranean termite Reticulitermes flaviceps (Isoptera:Rhinotermitidae). Conserv Genet Resour. 8:451–453.