Abstract
Cenchrus ciliaris is an important pasture resource for arid region and owing to apomixis, it has been very important for development and distribution of cultivars and agro-types. In present study complete chloroplast genome of C. ciliaris was sequenced. The size of chloroplast genome is 138737 bp length with overall GC content 38.6%. It exhibited regular quadripartite structure with 81053 bp of LSC region, 16108 bp of SSC region and 20788 bp of each IR region. A total of 130 genes were identified, including 87 coding genes, 32 tRNAs, 7 ribosomal RNAs and 4 pseudogenes. Phylogenetic analysis was performed using 13 other members representing major subfamilies of Poaceae.
Buffel grass (Cenchrus ciliaris) is perennial, drought tolerant, deep rooted grass and responds more rapidly to rain. It is native to Africa, India and Indonesia. Once it is established it can withstand heavy grazing, has high nutritional value, spreads rapidly and therefore extremely valuable pasture resource for arid ecosystems like desert where very few resources are available (Janice Citation2005; Marshall et al. Citation2012). Facultative apomixis reported in C. ciliaris which is important for the development and distribution of cultivars and agro-types (Bray Citation1978). Because of these characteristics it is considered to be a ‘wonder crop’ (Hanselka Citation1988).
C. ciliaris was collected from Kutchh desert (23.85°N 69.72°E) and deposited at Department of Biosciences, Saurashtra University under voucher number SUPG005. Fresh leaves were kept for 48 hours in 4 °C to decrease starch level. Chloroplast DNA isolation was performed according to (Shi et al. Citation2012) and checked on 1% agarose gel electrophoresis. This DNA was sequenced using high throughput ion torrent genome machine with ion torrent server (torrent suite v3.2). Reference guided assembly was performed using CLC (mapping parameters: Mismatch cost =2, Insertion cost =3, Deletion cost =3, length fraction= 0.5, similarity =0.8) with already available chloroplast genome of Cenchrus americanus (NC_024171) as the reference genome. Contigs with more than 50 × sequence depth were used for reference guided assembly. The vote majority conflict resolution mode of CLC was used in order to ensure inclusion of only chloroplast specific reads thus avoiding contribution of nuclear and mitochondrial reads to the consensus sequence. Reads were also de novo assembled using CLC. Consensus sequence derived from reference assembly was compared and corrected with de novo assembly. Plastome annotation was performed in CpGAVAS (Liu et al. Citation2012) and DOGMA (Wyman et al. Citation2004). Further tRNA genes were confirmed using tRNAscan-SE 2.0 (Lowe and Eddy Citation1997). Complete chloroplast genome of C. ciliaris was submitted to NCBI GenBank (accession no. MH286942).
Chloroplast genome length of C. ciliaris is 138737 bp with overall 38.6% GC content. It exhibited regular quadripartite structure with 81053 bp of LSC region, 16108 bp of SSC region and 20788 bp of each IR region. A total of 130 genes were identified, including 87 coding genes, 32 tRNAs, 7 ribosomal RNAs and 4 pseudogenes. Total 17 genes including 8 protein coding genes, 6 transfer RNA genes and 3 ribosomal RNA genes present in IR region are found to be duplicated.
Phylogenetic analysis was performed using complete chloroplast genome of C. ciliaris with other 13 members of Poaceae. As Cenchrus belongs to Panicoideae 4 members selected from this group and from each subfamily of Poaceae one important member was selected. Phylogenetic tree clearly separates Panicoideae group containing 3 Cenchrus species, Saccharum officinarum, Sorghum bicolor and Zea mays from other members with nearly 100% bootstrap support ().
Figure 1. Phylogenetic tree based on complete chloroplast genome sequence of Cenchrus ciliaris and other 13 members of Poaceae. Phylogenetic tree constructed using maximum likelihood method and numbers on the each node are bootstrap values based on 1000 replicates.
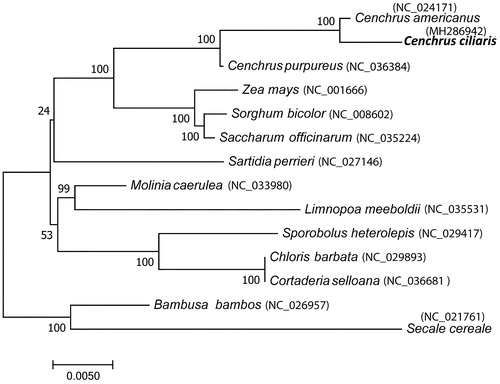
As it is difficult to distinguish C. ciliaris from closely related other Cenchrus species and complete chloroplast genome provide effective tool for accurate identification and phylogenetic analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bray RA. 1978. Evidence for facultative apomixis in Cenchrus Ciliaris. Euphytica. 27:801–804.
- Hanselka CW. 1988. Buffelgrass: south Texas wonder grass. Rangelands. 10:279–281.
- Janice J. 2005. Is there a relationship between herbaceous species richness and buffel grass (Cenchrus Ciliaris)? Austral Ecol. 30:505–517.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Marshall VM, Lewis MM, Ostendorf B. 2012. Buffel grass (Cenchrus Ciliaris) as an invader and threat to biodiversity in arid environments: A review. J Arid Environ. 78:1–12.
- Shi C, Hu N, Huang H, Gao J, Zhao Y-J, Gao L-Z. 2012. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. Plos ONE. 7:e31468
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
