Abstract
The first complete chloroplast genome sequences of Korean endemic Epilobium in Ulleung Island, Epilobium ulleungensis, were reported in this study. The E. ulleungensis plastome was 160,912 bp long, with the large single copy (LSC) region of 88,915 bp, the small single copy (SSC) region of 17,327 bp, and two inverted repeat (IR) regions of 27,335 bp. The plastome contained 131 genes, including 84 protein-coding, eight ribosomal RNA, and 37 transfer RNA genes. The overall GC content was 36.5%. Phylogenetic analysis of nine representative plastomes within the family Onagraceae suggests strongly that E. ulleungensis is sister to the clade containing species of Oenothera in tribe Onagreae.
Owing to its remarkable diversity in morphology, ecology, and cytology, the genus Epilobium L., with approximately 175 species, represents the largest group in Onagraceae and is mainly distributed in temperate regions (Raven Citation1988; Hoch and Raven Citation1992; Baum et al. Citation1994). As for infrageneric classification system, eight highly distinctive sections have been proposed and recent molecular phylogenetic study recognized two main clades, i.e. sect. Epilobium and the ‘xerophytic’ clade (six sections included), and sect. Chamaenerion being the earliest diverged lineage within the genus (Raven Citation197Citation6, Citation1988; Baum et al. Citation1994). Of eight sections delimited based on morphological and cytological characters (Raven Citation1976), sect. Eplilobium is the largest one (ca. 150 spp.) with major radiation in Australasia and smaller ones in temperate South America and southern Africa (Raven and Raven Citation1976; Baum et al. Citation1994). Although preliminary nrDNA ITS phylogeny within the genus was performed and infrageneric classification system was evaluated (Baum et al. Citation1994), several questions (e.g. relationship within the ‘xerophytic’ clade and sectional relationship among Boisduvalia, Zauschneria, and Currania) require additional study based on extensive sampling within the genus and additional plastid and independent nuclear markers. In Korea, a total of 12 species of Epilobium are currently recognized (National List of Species of Korea Citation2018), including one recently described insular endemic E. ulleungensis on Ulleung Island (Chung et al. Citation2017). Neither phylogenetic relationships nor complete chloroplast genome sequences is available among species of Epilobium in Korea. Therefore, in this study, we sequenced the plastome of E. ulleungensis and compared it to other plastomes in the family Onagraceae.
Total DNA (Voucher specimen: JMC15101, KH) was isolated using the DNeasy plant Mini Kit (Quiagen, Carlsbad, CA) and sequenced by the Illumina HiSeq 4000 (Illumina Inc., San Diego, CA). A total of 11,439,577 paired-end reads were obtained and assembled de novo with Velvet v. 1.2.10 using multiple k-mers (Zerbino and Birney Citation2008). The tRNAs were confirmed using tRNAsacn-SE (Lowe and Eddy Citation1997). The total plastome length of E. ulleungensis (MH198310) was 160,912 bp, with large single copy (LSC; 88,915 bp), small single copy (SSC; 17,327 bp), and two inverted repeats (IRa and IRb; 27,335 bp each). The overall GC content was 38.2% (LSC, 36.3%; SSC, 33.2%; IRs, 42.8%) and the plastome contained 131 genes, including 84 protein-coding, eight rRNA, and 37 tRNA genes. A total of 18 genes were duplicated in the inverted repeat regions, including seven tRNA, four rRNA, and seven protein coding genes including ndhF gene. The complete ycf1 gene was located in SSC and the infA gene located in LSC became a pseudogene.
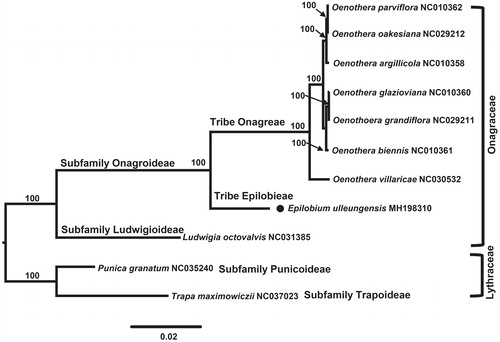
To confirm the phylogenetic position of E. ulleungensis, nine representative plastomes of Onagraceae and two outgroup species from Lythraceae were aligned using MAFFT v.7 (Katoh and Standley Citation2013) and maximum likelihood (ML) analysis was conducted based on the concatenated 77 coding genes using IQ-TREE v.1.4.2 (Nguyen et al. Citation2015). The ML tree () strongly suggested that E. ulleungensis (tribe Epilobieae) is sister to the clade containing species of Oenothera (tribe Onagreae).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Baum DA, Sytsma KJ, Hoch PC. 1994. A phylogenetic analysis of Epilobium (Onagraceae) based on nuclear ribosomal DNA sequences. Syst Bot. 19:363–388.
- Chung J-M, Shin J-K, Sun E-M, Kim H-W. 2017. A new species of Epilobium (Onagraceae) from Ulleungdo Island, Korea, Epilobium ulleungensis. Korean J Pl Taxon. 47:100–105.
- Hoch PC, Raven PH. 1992. Boisduvalia, a coma-less Epilobium (Onagraceae). Phytologia. 73:456–459.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- National List of Species of Korea. 2018. National Institute of Biological Resources. [accessed 2018 May 2]. http://kbr.go.kr
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Watts GB, Gard M. Md. 1937. Generic and sectional delimitation in Onagraceae, tribe Epilobieae. Ann Mo Bot Gard. 11:326–340.
- Raven PH. 1988. Onagraceae as a model of plan evolution. In: Gottlieb LD, Jain SK, editors. Plant evolutionary biology. New York (NY): Chapman and Hall Press; p. 85–107.
- Raven PH, Raven TE. 1976. The genus Epilobium in Australia: a systematic and evolutionary study. Dept Sci Industr Res Bull. 216:1–321.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.