Abstract
Scuticociliates are dangerous parasitic pathogens for in worldwide mariculture. Scuticociliates cause high mortality to marine fish. After an outbreak of scuticociliatosis in Takifugu rubripes in Liaoning Province, northern China, Uronema marinum, a scuticociliate, was identified. In this study, using Illumina MiSeq next-generation sequencing, the mitochondrial genome of U. marinum was assembled and analysed phylogenetically using mitochondrial genomes of other scuticociliates. The complete U. marinum mitochondrial genome was 39,845 bp; it contained two rRNAs, six tRNAs, and 39 protein-coding genes (PCGs). From the 39 PCGs, 15 PCGs were located on the heavy strand, and 24 PCGs on the light strand of U. marinum mitogenome. The phylogenetic tree showed that there were two main clades, Oligohymenophorea and Spirotrichea. Nine ciliate species were clustered together within Oligohymenophorea; Uronema marinum was a separate cluster sharing a relatively close ancestry with Hymenostomatida. The results of this study will help advance the systematics, and studies of evolution and molecular epidemiology of scuticociliates.
Uronema marinum is a scuticocilaite that belongs to order Shieldophilus, family Sphingidae, and genus Uronema. It causes scuticociliatosis, a disease with a high mortality in marine fish (Yoshinaga and Nakazoe, Citation1993; Sterud et al. Citation2000; Jee et al. Citation2001; Anderson et al. Citation2009). Recently, after an outbreak of severe scuticociliatosis in Takifugu rubripes in factory farming in Liaoning Province, northern China (38.9728 N and 121.3326 E), we isolated a pure pathogenic ciliate from the skin ulcers of ill T. rubripes and identified it as the scuticociliate U. marinum. Although there are many scuticociliate species reported, such as Pseudocohnilembus persalinus (Kim et al. Citation2004), Phiasterides dicentrachi (Sungmi et al. Citation2004), and Miamiensis avidus (Jung et al. Citation2005), the information of Scuticociliatida mitochondrial genome is very limited. Moreover, the mt genome sequence of U. marinum was still in the blank.
In this study, the total DNA of U. marinum was extracted, and stored in Dalian Key Laboratory of Marine Animal Disease Control and Prevention, Dalian Ocean University, Dalian, China. The mitochondrial genome of U. marinum was assembled using Illumina MiSeq Next-generation sequencing by SC Gene Company (Guangzhou, China). The sequence of U. marinum mitochondrial genome was deposited in GenBank (accession number MG272262). The whole mitogenome of U. marinum was 39,845 bp. Its nucleotide composition was as follows: A, 39.83%; T, 41.17%; C, 8.94%; and G, 10.06%. There were two rRNAs, six tRNAs, and 39 protein-coding genes (PCGs). Furthermore, the genes were coded on both the heavy strand and the light strand. From the 39 PCGs of U. marinum mitogenome, 15 PCGs were located on the heavy strand, and 24 PCGs on the light strand. Six genes (cox1, nad1_a, orf346, rpl2, ymf65, cob) used the start codon ATT. Seventeen genes (rpl14, ymf70, nad4, rps3, nad10, nad7, rps14, rpl6, ymf75, nad1_b, atp9, rpl16, nad3, nad9, cox2, ymf56, ymf68) used the start codon ATG. Eight genes (ymf57, rps13, nad2, yejR, orf202, nad4l, nad5, ymf67) used the start codon ATA. Two genes (nad6, rps19) used the start codon GTG. Four genes (orf141, rps12, ymf64, ymf63) used the start codon TTA. The genes orf149 and ymf66 used the start codon TTG and ATC, respectively. The stop codon TAA was present in 38 PCGs. Only the gene ymf56 used the stop codon TAG. The G + C content of PCGs was 18.4%, very close to the 19% average G + C content of U. marinum mitogenome. The overall number of codon occurrences was 10,345. The most frequent amino-acid encoding codons were Leu (13.71%), Phe (11.69%), Asn (9.98%), Ile (9.73%), Lys (8.08%), Tyr (7.51%), Ser (6.83%), and Val (5.43%). Two rRNA genes (rnl, 2,566 bp; and rns 1,509 bp) were located on the heavy strand of U. marinum mitogenome. The G + C content of rRNA genes was 23.38% for rnl, and 24.78% for rns, which was higher than the average 19% G + C content of the U. marinum mitogenome. There were six tRNAs on the mitogenome of U. marinum. tRNA-Glu, tRNA-Met, and tRNA-Trp were on the heavy strand, and tRNA-Tyr, tRNA-Phe, and tRNA-His were on the light strand. These six tRNA genes had the predicted typical cloverleaf secondary structure.
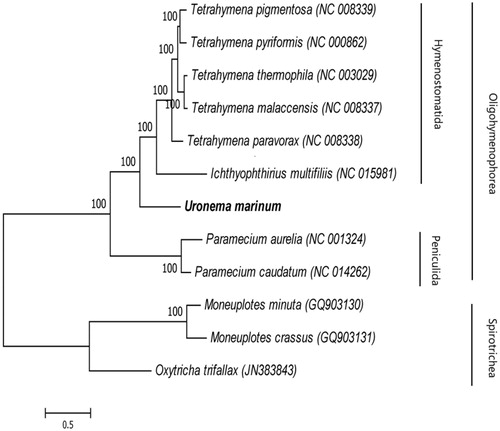
Phylogenetic relationships between 12 ciliates in the Oligohymenophorea taxa were analysed according to the nucleotide and amino-acid sequences of 13 PCGs from GenBank using BI and ML methods. The resulting phylogenetic tree showed that there were two main clades, Oligohymenophorea and Spirotrichea (). Nine ciliate species were clustered together within Oligohymenophorea; Uronema marinum was a separate cluster, sharing a relatively close ancestry with Hymenostomatida.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Anderson SA, Hulston DA, Mcveagh SM, Webb VL, Smith PJ. 2009. In vitro culture and cryopreservation of Uronema marinum isolated from farmed New Zealand groper (Polyprion oxygeneios). J Microbiol Methods. 79:62–66.
- Jee BY, Kim YC, Park MS. 2001. Morphology and biology of parasite responsible for scuticociliatosis of cultured olive flounder Paralichthys olivaceus. Dis Aquat Org. 47:49–55.
- Jung SJ, Kitamura S, Song JY, Joung IY, Oh MJ. 2005. Complete small subunit rRNA gene sequence of the scuticociliate Miamiensis avidus pathogenic to olive flounder Paralichthys olivaceus. Dis Aquat Org. 64:159–162.
- Kim SM, Cho JB, Kim SK, Nam YK, Kim KH. 2004. Occurrence of scuticociliatosis in olive flounder Paralichthys olivaceus by Phiasterides dicentrarchi (Ciliophora: Scuticociliatida). Dis Aquat Org. 62:233–238.
- Kim SM, Cho JB, Lee EH, Kwon SR, Kim SK, Nam YK, Kim KH. 2004. Pseudocohnilembus persalinus (Ciliophora: Scuticociitida) is an additional species causing scuticociliatosis in olive flounder Paralichthys olivaceus. Dis Aquat Org. 62:239–243.
- Sterud E, Hansen MK, Mo TA. 2000. Systemic infection with Uronema-like ciliates in farmed turbot, Scophthalmus maximus (L.). J Fish Dis. 23:33–37.
- Sungmi K, Jaebum C, Sungkoo K, Yoonkwon N, Kihong K. 2004. Occurrence of scuticociliatosis in olive flounder Paralichthys olivaceus by Phiasterides dicentrarchi (Ciliophora: Scuticociliatida). Dis Aquat Org. 62:233–238.
- Yoshinaga T, Nakazoe JI. 1993. Isolation and in vitro Cultivation of an Unidentified Ciliate Causing Scuticociliatosis in Japanese Flounder (Paralichthys olivaceus). Fish Pathol. 28:131–134.