Abstract
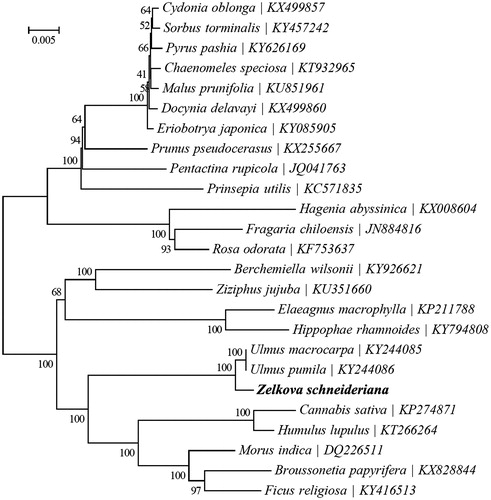
Zelkova schneideriana Hand.-Mazz. (Ulmaceae) is an endangered species endemic to China. In this study, we reported its complete chloroplast (cp) genome based on Illumina pair-end sequencing. The whole genome was 158,999 bp long, consisting of a pair of inverted repeat (IR) regions of 26,427 bp, a large single copy (LSC) region of 87,397 bp and a small single copy (SSC) region of 18,748 bp. The cp genome contained 133 genes, including 88 protein-coding genes (80 PCG species), 37 tRNA genes (30 tRNA species), and eight rRNA genes (4 rRNA species). The overall G+C content of the whole genome was 35.6%, and the corresponding values of the LSC, SSC, and IR regions were 33.0, 28.3, and 42.4%, respectively. The maximum likelihood phylogenetic analysis of 25 selected chloroplast genomes demonstrated that Z. schneideriana was closely related to Ulmus macrocarpa and Ulmus pumila.
Zelkova schneideriana Hand.-Mazz., belonging to family Ulmaceae, is mainly distributed in Huai River Basin, Qinling Mountains, and the Lower-middle Reaches of the Yangtze River in China (Zhang et al. Citation2013). As a consequence of the excessively cut for utilizing, this rare species endemic to China is under the threat of extinction and has been classified as a vulnerably Endangered plant listed into Chinese Dangerous Plant Red Paper (Class II) (Fu Citation1992). Therefore, there is immediate need to take effective measures to protect this species. Here, we characterized the complete chloroplast (cp) genome of Z. schneideriana based on high-throughput Illumina sequencing technology, which will contribute to the further studies on the genetic diversity, systematic evolution, and conservation of this Endangered tree species. The annotated cp genome of Z. schneideriana has been deposited into GenBank with the accession number MG717940.
Fresh leaves of a Z. schneideriana individual were collected from Fujian Agriculture and Forestry University (Fujian, China; 26°03′N, 119°16′E). A shotgun library was constructed and whole genome sequencing was performed with 150 bp pair-end (PE) reads on the Illumina Hiseq 2500 platform (Illumina, CA). In total, 30,214,272 raw reads of Z. schneideriana were obtained and their qualities were filtered and trimmed using NGS QC Toolkit v2.3.3 (Patel and Jain Citation2012). Then, 144,092 clean reads were assembled by MITObim v1.8 (Hahn et al. Citation2013) with the sequences of published Ulmus davidiana (KY244082) cp genomes as the reference. The cp genome sequence was annotated using the program Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand).
The complete cp genome of Z. schneideriana was 158,999 bp in length, containing a pair of inverted repeat (IR) regions of 26,427 bp which were separated by a large single copy (LSC) region of 87,397 bp and a small single copy (SSC) region of 18,748 bp. It encoded 133 genes, including 88 protein-coding genes (80 PCG species), 37 transfer RNA genes (30 tRNA species), and eight ribosomal RNA genes (four RNA species). Among these genes, a total of 19 gene species had two copies in the IR regions, including eight PCG species (rps19, rpl2, rpl23, ycf2, ycf15, ndhB, rps7, and ycf1), seven tRNA species (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU), and four rRNA species (rrn16, rrn23, rrn4.5, and rrn5). Nine PCG species (rps16, rpoC1, rpl12, petB, petD, rpl16, rpl2, ndhB, and ndhA) and six tRNA species (trnK-UUU, trnG-GCC, trnL-UAA, trnV-UAC, trnI-GAU, and trnA-UGC) possessed a single intron, while another three gene species (rps12, ycf3, and clpP) contained two introns. The overall GC content of the cp genome is 35.6%, with the corresponding values of 33.0, 28.3, and 42.4% for the LSC, SSC, and IR regions, respectively.
To ascertain its phylogenetic position within the order Rosales, a maximum likelihood (ML) tree was reconstructed using the GTR + G + I model, with 1000 bootstrap replicates, in RAxML v.8.2.8 (Stamatakis Citation2014). The phylogenetic analysis indicated that Z. schneideriana was closely related to Ulmus macrocarpa and Ulmus pumila ().
Acknowledgements
We thank Dr Yafu Zhou for linguistic comments.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Fu LK. 1992. China plant red data book-rare and endangered plants. Beijing: Science Press.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Zhang RQ, Liu HL, Wang LD, Jin XL. 2013. Aseptic seeding techniques in Zelkova schneideriana. Nonwood Forest Res. 1:139–142.