Abstract
Cucurbita pepo is an important economic plant cultivated widely in the world. The complete chloroplast genome sequence of C. pepo is reported here. The genome is 157,343 bp in length and exhibits a typical quadripartite structure of the large (LSC, 87,970 bp) and small (SSC, 18,167 bp) single-copy regions, separated by a pair of inverted repeats (IRs, 25,603 bp). A total of 131 genes were predicted including 85 protein-coding genes, eight rRNA genes and 38 tRNA genes. Further, phylogenetic analysis showed that C. pepo were closely related to other species in the family Cucurbitaceae. The complete chloroplast genome of C. pepo would be taken as a useful molecular tool for species discrimination, taxonomy, and phylogenetic relationships in the family Cucurbitaceae.
Cucurbita pepo L. is a biologically and economically important crop (Montero-Pau et al. Citation2017), commonly known as ‘Zucchini’ or ‘Summer squash’. The species with high genetic diversity is a world-wide cultivated vegetable of American origin (Formisano et al. Citation2012). Chloroplast genome is maternally inherited in C. pepo, and considered a useful molecular tool to discriminate species, explore species divergence, and trace biogeographic history (Lim et al. Citation1990; Schaefer et al. Citation2009; Logan et al. Citation2015). Moreover, the pericarp pigment would be regulated by the different expression of genes in the plastid genome to show different skin color (Schaffer et al. Citation1984) and the cotyledon senescence has been delayed by the expression of plastid-encoded genes (Mishev et al. Citation2011). In this study, we characterized the complete chloroplast genome sequence of Cucurbita pepo for promoting the studies on the phylogenetic relationships, germplasm exploration, genetic breeding, and physiological mechanism.
The genomic DNA was isolated from the leaves of C. pepo subsp. pepo (Accession: BGV004370) using the CTAB method as previously described (Montero-Pau et al. Citation2017). Genomic DNA was subjected to construct a 500 bp pair-end library and sequenced by illumina HiSeq2000 (Macrogen, Seoul, Republic of Korea). The whole genome sequence data have been deposited for academic use in the SRA database of NCBI with the accession (SRR5581782), it was downloaded and trimmed, high quality pair-end reads (2 × 101 bp) of 0.23 Gb (∼0.8× coverage to nuclear genome) were randomly extracted using Seqtk and assembled with using the Plasmidspades.py in SPAdes (v3.10.1) (Bankevich et al. Citation2012). Contigs representing the chloroplast genome were retrieved, ordered and joined into a single draft sequence by comparison with the chloroplast genome of Cucumis melo (GenBank accession no. NC_015983.1) as a reference (Rodriguez-Moreno et al. Citation2011). The gaps in the chloroplast single draft sequence of were closed by using GapCloser (v1.12-r6). The draft sequence was then confirmed and manually corrected by PE read mapping. Finally, the draft sequence was annotated using the two integrated web servers, CpGAVAS (Chang et al. Citation2012) and DOGMA (Wyman et al. Citation2004), and manually corrected by visual inspection using IGV (Robinson et al. Citation2011).
A typical chloroplast genome consists of four distinct regions, a large and a small single copy region (LSC and SSC, respectively) separated by two inverted repeat regions (IRa and IRb). The complete chloroplast genome of Cucurbita pepo (GenBank accession number MH031787) is 157,343 bp in length with 37.16% GC contents, and exhibits a typical quadripartite structure, consisting of a pair of IRs (25,603 bp) separated by the LSC (87,970 bp) and SSC (18,167 bp) regions. There is a total of 131 genes, including 85 protein-coding genes, eight rRNA genes and 38 tRNA genes; six of the protein-coding genes, six of the tRNA genes and four rRNA genes are duplicated within the IRs.
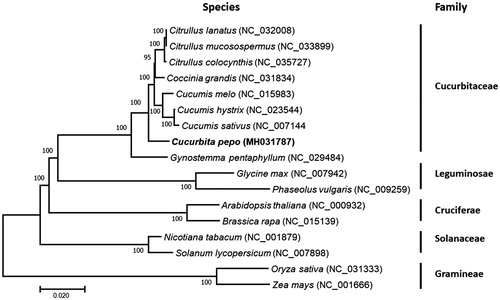
To determine the phylogenetic position of C. pepo, a phylogenetic analysis was carried out among 17 complete chloroplast genomes that are derived from Cucurbitaceae, Leguminosae, Cruciferae, Solanaceae, and Gramineae. The phylogenetic tree was constructed by maximum-likelihood method using the program PhyML in Unipro UGENE v1.29.0 (Okonechnikov et al. Citation2012). The results showed that C. pepo was clustered into the family Cucurbitaceae and closer to Gynostemma than Citrullus, Coccinia, and Cucumis (). In addition, other species were also well clustered into the clades corresponding to their families.
Figure 1. Phylogenetic tree showing relationship between C. pepo and other 16 species belonging to different families. Phylogenetic tree was constructed based on the complete chloroplast genomes using maximum likelihood (ML) with 1000 bootstrap replicates. Numbers in each the node indicated the bootstrap support values.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Chang L, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Formisano G, Roig C, Esteras C, Ercolano MR, Nuez F, Monforte AJ, Picó MB. 2012. Genetic diversity of Spanish Cucurbita pepo landraces: an unexploited resource for summer squash breeding. Genet. Genet Resour Crop Evol. 59:1169–1184.
- Lim H, Gounaris I, Hardison RC, Boyer CD. 1990. Restriction site and genetic map of Cucurbita pepo chloroplast DNA. Curr Genet. 18:273.
- Logan K, Newsom LA, Ryan TM, Clarke AC, Smith BD, Perry GH. 2015. Gourds and squashes (Cucurbita spp.) adapted to megafaunal extinction and ecological anachronism through domestication. P Natl Acad Sci USA. 112:15107.
- Mishev K, Dimitrova A, Ananiev ED. 2011. Darkness affects differentially the expression of plastid-encoded genes and delays the senescence-induced down-regulation of chloroplast transcription in cotyledons of Cucurbita pepo L. (Zucchini). Z Naturforsch C. 66:0159–0166.
- Montero-Pau J, Blanca J, Bombarely A, Ziarsolo P, Esteras C, Martí-Gómez C, Ferriol M, Gómez P, Jamilena M, Mueller L, et al. 2017. De novo assembly of the zucchini genome reveals a whole genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol J. 16:1161–1171.
- Okonechnikov K, Golosova O, Fursov M. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 28:1166–1167.
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol. 29:24–26.
- Rodriguez-Moreno L, Gonzalez VM, Benjak A, Marti MC, Puigdomenech P, Aranda MA, Garcia-Mas J. 2011. Determination of the melon chloroplast and mitochondrial genome sequences reveals that the largest reported mitochondrial genome in plants contains a significant amount of DNA having a nuclear origin. BMC Genomics. 12:424.
- Schaefer H, Heibl C, Renner SS. 2009. Gourds afloat: a dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc Roy Soc B-Biol Sci. 276:843–851.
- Schaffer AA, Boyer CD, Gianfagna T. 1984. Genetic control of plastid carotenoids and transformation in the skin of Cucurbita pepo L. fruit. Theor Appl Genet. 68:493–501.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
