Abstract
In this study, the complete mitochondrial genome of Chionodraco rastrospinosus was obtained, which was 17598 bp including 2 ribosomal RNAs, 13 protein-coding genes, 22 transfer RNAs, and a non-coding control region. The length of D-loop was 1332 bp and its contents of A, T, C, and G were 30.3%, 27.6%, 26.8%, and 15.3%. The complete mtDNA sequences of C. rastrospinosus and other 14 species were used to reconstruct the phylogenetic tree, suggested that C. rastrospinosus was closest to two species of Chionodraco. The study would provide a basic data for further research on population structure, conservation genetics and molecular evolution of C. rastrospinosus.
Chionodraco rastrospinosus is widely distributed in the South Shetland Islands, the South Orkney Islands, and the Antarctic Peninsula (Kock K-H Citation1993). In these areas, it is found most commonly at depths from 200–400 m where the temperature is close to –1 °C. Because of genetic mutations, they completely lack the oxygen-binding haemoglobin and the colour of blood is translucent instead of red (Kock K-H Citation2005). Papetti reported the partial sequence of C. rastrospinosus and no ND6 gene (Papetti et al. Citation2007). However, the current study found that Chionodraco hamatus (Song et al. Citation2016), which also belonged to Chionodraco, had ND6 gene, so we did the research. Besides, the complete mitochondrial genome sequence of C. rastrospinosus would be useful for understanding population structure, molecular systematic, and conservation genetics.
Adult fish of C. rastrospinosus was collected from Antarctic (62°52′54″S, 59°28′54″W), it was transported to East China Sea Fisheries Research Institute Chinese Academy of Fishery Science after freezing at –80 °C. The genomic DNA was extracted from muscle tissues and sequenced using the Illumina HiSeq2000 platform (Illumina, San Diego, CA). The primers for amplification were designed according to the sequence of C. hamatus (KT921282). We obtained the complete mitogenome and submitted it to Genbank database with an accession number MF622064.
It was a circular molecule of 17598 bp in length, including 2 rRNAs, 13 protein-coding genes, 22 tRNAs, and a non-coding control region. The contents of A, T, C, and G were 26.42%, 26.10%, 30.05%, and 17.41%, respectively. Among the 22 tRNAs, 8 tRNAs (tRNAPro, tRNAGlu, tRNASer, tRNATyr, tRNACys, tRNAAsn, tRNAAla, and tRNAGln) were encoded by L-strand, when others were encoded by H-strand. In 13 protein-coding genes, two kinds of start codons were identified, two (CO1 and ATP6) were started with GTG, whereas others started with ATG. Five genes (ND1, CO1, ATP8, ATP6, and ND4L) ended with TAA or TAG and the others ended with T or TA. The length of D-loop was 1332 bp, it was shorter than C. hamatus and its overall base composition was 30.3% for A, 27.6% for T, 26.8% for C, and 15.3% for G, with a high A + T content of 57.9%, accord with the structural characteristics of AT rich.
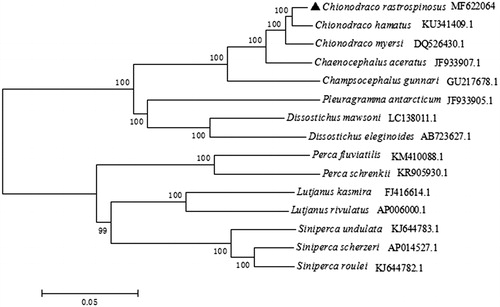
Phylogenetic analysis was performed using the Neighbour-joining method in MEGA 5.0 based on complete mitogenome of C. rastrospinosus and other 14 species (). The phylogenetic tree showed that C. rastrospinosus clustered with C. hamatus and C. myersi and then together with other two fishes in family Channichthyidae. The result was similar to the outcome of Song’s research on C. hamatus (Song et al. Citation2016).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kock K-H. 1993. Antarctic fish and fisheries. J Mar Biol Assoc UK. 73:252–359.
- Kock K-H. 2005. Antarctic icefishes (Channichthyidae): a unique family of fishes. A review, Part II. Polar Biol. 28:862–895.
- Papetti C, Liò P, Rüber L, Patarnello T, Zardoya R. 2007. Antarctic fish mitochondrial genomes lack ND6 gene. J Mol Evol. 65:519–528.
- Song W, Li LZ, Huang HL, Meng YY, Jiang KJ, Zhang FY, Chen XZ, Ma LB. 2016. The complete mitochondrial genome of Chionodraco hamatus (Notothenioidei: Channichthyidae) with phylogenetic consideration. Mitochondrial DNA Part B. 1:52–53.