Abstract
The complete mitochondrial genome of Protosalanx chinensis was determined to be 16,879 bp in length. It consists of 13 protein-coding genes, 22 tRNAs, 2 ribosomal RNAs, and a control region. The gene composition and the structural arrangement of the P. chinensis complete mtDNA were identical to most of the other vertebrates. The molecular data here we presented could play a useful role to study the evolutionary relationships and population genetics of Salangidae fish.
The clearhead icefish Protosalanx chinensis is a small, pelagic, carnivorous species, which mainly distributes in the coastal area of the East China Sea, Yellow Sea, and Bohai Sea and their adjacent rivers and lakes in eastern Asia (Saruwatari et al. Citation2002). Because of its high commercial value, P. chinensis has been traditionally exploited in China and are also one of the most important exported fish to European, Asian and American markets (Armani et al. Citation2011). To facilitate the future researches of taxonomic resolution, population genetic structure and phylogeography, the complete mitochondrial genome of P. chinensis was determined (GenBank number MH330683).
The samples were collected from the Taihu Lake, Wuxi city, China (31°24′44″ N, 120°19′43″ E). Muscle samples were fixed in 95% ethanol and preserved in Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences. Twenty-eight pairs of PCR primers were designed to based on sequences from related fish Neosalanx tangkahkeii and N. anderssoni. The complete mitochondrial genome of P. chinensis is 16,879 bp in length. The overall nucleotide composition was A (25.5%), T (25.1%), C (31.4%), and G (18%), without a significant AT bias of 50.6%. Mitogenome database were annotated by MitoFish (Iwasaki et al. Citation2013). It consists of 13 typical vertebrate protein-coding genes, 22 tRNA, 2 rRNA genes and 1 control region. ND6 gene and 8 tRNA genes were encoded on the light (L) strain; the remaining genes were encoded on the heavy (H) strain. For protein-coding genes, 12 were initiated with the orthodox ATG except for COI with GTG, and they had 4 types of stop codons (TAA, TAG, TA–, and T–). All tRNAs were recognized by tRNAscan-SE1.21 (Lowe and Eddy Citation1997), and their length ranged from 65 to 75 bp. The 2 rRNA genes contained 943 and 1710 bp, respectively. The control region of the P. chinensis mitogenome was 1221 bp in length, which was slightly longer than that of N. tangkahkeii and N. anderssoni, as it showed three tandem repeat units.
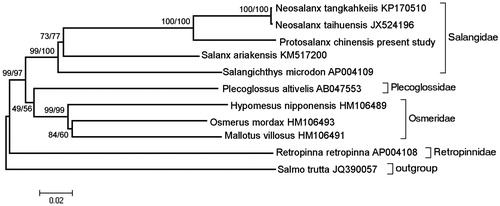
Phylogenetic analysis was performed by MEGA 6.06 (Tamura et al. Citation2013) based on complete mitogenome sequence of P. chinensis and those of 10 closely related species using maximum parsimony (MP) and neighbour-joining (NJ) method with 1000 bootstrap replicates (). Identical tree topologies were recovered from the MP and NJ analyses, and the only differences were bootstrap values. As indicated by the tree, different species from the same family clustered together (e.g. Salangidae and Osmeridae), and species from Osmeroidei formed a monophyletic group while Salmonidae positioned as an outgroup.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Armani A, Castigliego L, Tinacci L, Gianfaldoni D, Guidi A. 2011. Molecular characterization of icefish, (salangidae family), using direct sequencing of mitochondrial cytochrome b gene. Food Control. 22:888–895.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, Nishida M. 2013. Mitofish and mitoannotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Saruwatari T, Oohara I, Kobayashi T. 2002. Salangid fishes: their past, present and future. Fish Sci. 68:71–74.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. Mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.