Abstract
Pyrrosia bonii is an important ornamental and medical fern from Polypodiaceae. Its complete chloroplast genome sequence was characterized through de novo assembly with Illumina sequencing data. The genome size is 158,174 bp, with large single copy (LSC, 82,479 bp) and small single copy (SSC, 21,723 bp) regions separated by a pair of inverted repeats (IR, 26,986 bp). A total of 132 genes were identified, including 88 protein coding genes, 35 tRNA genes, eight rRNA genes, and one pseudogene. Maximum-likelihood phylogenetic tree revealed that P. bonii was most closely related to Lepisorus clathratus. The study will greatly facilitate the chloroplast phylogenomics of Polypodiaceae.
Pyrrosia bonii (Christ ex Giesenh.) Ching is an epiphytic tropical fern belonging to Polypodiaceae. It has two key characters of this genus, i.e. stellate hairs and connective venation pattern (Zhang et al. Citation2013). As a terrestrial fern, the species mainly grows on rocks in forest at an altitude of 300–1100 m with distribution in China including Guangxi and Guizhou, as well as Vietnam. Pyrrosia bonii has not only unique ornamental value, but also high medical value. The dried leaves of P. bonii have been used to substitute for medicinal materials ‘Shiwei’ to treat urinary infection and urolithiasis, although it has not been recorded in the Chinese pharmacopoeia (National Pharmacopoeia Committee Citation2010). In addition, there are many difficulties and controversies in species-level classification in Pyrrosia due to high similarity in morphology (Wei et al. Citation2017). The monophyly and systematic position of Pyrrosia in Platycerioideae also occur controversy (Hovenkamp Citation1986). Hence, sequencing of complete chloroplast genome of P. bonii will be conducive to a deeper and better understanding of classification and phylogeny of Pyrrosia.
Total genomic DNA was extracted from the fresh leaves of P. bonii using the Tiangen Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China), which were collected from South China Botanical Garden, Chinese Academy of Sciences (23°11′3.56″N, 113°21′43.28″E). The specimen was saved in Herbarium of Sun Yat-sen University (SYS; voucher: SS Liu 201615). After DNA was broken by Covaris M220 (Covaris Inc., Woburn, MS), an average 300 bp Illumina library was constructed and sequenced on the Illumina Hiseq 2500 platform (Illumina, San Diego, CA). We separately used Trimmomatic v0.32 (Bolger et al. Citation2014) and FastQC v0.10.0 (Andrews Citation2010) to trim reads and visualize the quality of the clean reads. A total of 1.86 G clean data was further assembled into the complete chloroplast genome using Velvet v1.2.07 (Zerbino and Birney Citation2008), which was annotated through DOGMA (Wyman et al. Citation2004) and tRNAscan-SE (Schattner et al. Citation2005). After we aligned complete chloroplast genome sequences of 11 ferns and Alsophila spinulosa as an outgroup using MAFFT v7.394 (Kazutaka and Standley Citation2013), phylogenetic relationships were presented by RAxML v8.2.10 (Stamatakis Citation2014) based on the maximum-likelihood method with 1000 bootstrap replicates.
Figure 1. Using RAxML v8.2.10, ML phylogenetic tree was constructed based on complete chloroplast genome sequences of 11 ferns and Alsophila spinulosa as an outgroup. The numbers in the nodes indicated the support values with 1000 bootstrap replicates.
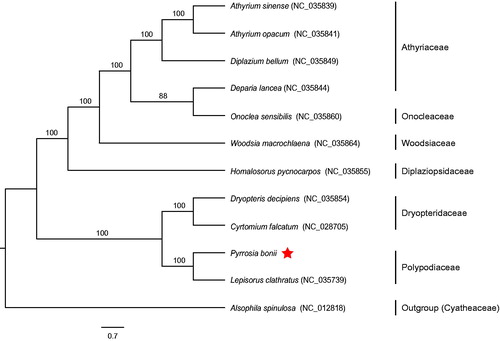
The complete chloroplast genome of P. bonii (GenBank accession no. MH352390) is a circular DNA molecule of 158,174 bp in length with a typical quadripartite structure including a large single copy (LSC) region (82,479 bp), a small single copy (SSC) region (21,723 bp) and a pair of inverted repeats (IRs) regions (26,986 bp). We annotated 132 genes, consisting of 88 protein coding genes (PCGs), 35 tRNA genes, eight rRNA genes and one pseudogene (ndhB). Among these genes, 15 genes (ndhB, rps16, atpF, rpoC1, petB, petD, ndhA, rpl16, rpl2, trnG-UCC, trnV-UAC, trnA-UGC, trnI-GAU, trnL-UAA, and trnT-UGU) contain one intron, while three genes (ycf3, clpP, and rps12) possess two introns. Fourteen genes are duplicated in two IR regions, involving in four PCGs (rps12, rps7, psbA, and ycf2), six tRNA genes (trnN-GUU, trnH-GUG, trnI-GAU, trnA-UGC, trnT-UGU, and trnR-ACG), and four rRNA genes (rrn4.5, rrn5, rrn16, and rrn23). The overall GC contents of chloroplast genome are 41.6%. ML tree reveals that P. bonii was most closely related to Lepisorus clathratus with 100% support value (). Our study will greatly facilitate the chloroplast phylogenomics of Polypodiaceae.
Disclosure statement
The authors declare no conflict of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Cambridge (UK): The Babraham Institute. [accessed 2017 July 20]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Hovenkamp PH. 1986. A monograph of the fern genus Pyrrosia (Polypodiaceae). Leiden Bot Ser. 9:1–280.
- Kazutaka K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- National Pharmacopoeia Committee. 2010. Pharmacopoeia of People’s Republic of China. Beijing: Chemical Industry Press. [in Chinese].
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–689.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Wei XP, Qi YD, Zhang XC, Li L, Hui Shang, Ran Wei, Liu HT, Zhang BG. 2017. Phylogeny, historical biogeography and characters evolution of the drought resistant fern Pyrrosia Mirbel (Polypodiaceae) inferred from plastid and nuclear markers. Sci Rep. 7:12757.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang XC, Lu SG, Lin YX, Qi XP, Moore S, Xing FW, Wang FG, Hovenkamp PH, Gilbert MG, Nooteboom HP, et al. 2013. Polypodiaceae. In: Wu ZY, Raven PH, Hong DY, eds., Flora of China. Vols. 2–3 (Pteridophytes). Beijing: Science Press; p. 758–850.
