Abstract
Salvia chanryoenica is an endemic species, which locates on the ridges of mountains in South Korea. In this study, we determined the complete chloroplast (cp) genome sequence of S. chanryoenica; cp genome of S. chanryoenica is 151,689 bp in length and consists of a large (82,903 bp) and small (17,634 bp) single-copy regions, separated by a pair of identical inverted repeats (25,576 bp). This genome contains unique 79 protein-coding genes, 30 tRNA, and 4 rRNA. The gene order and organization of the S. chanryoenica are consistent with those of other Lamiaceae cp genomes. The overall GC content of the whole genome was 37.9%. Phylogenetic tree constructed based on 71 protein-coding genes demonstrated a sister relationship within genus Salvia.
The genus Salvia L. is a member of Lamiaceae, which comprises approximately 900 species with a worldwide distribution. It also has economic and medicinal value known as oriental medicine in Central Asia (Zhou et al. Citation2005; Zhong et al. Citation2009). Three species of Salvia (S. plebeia, S. japonica, and S. chanroenica) and classification of the genus have been reported from Korea. Based on the life cycle, morphological characters such as the number of leaves, shapes, and hair of plants (Donoghue Citation2002) are determined. Salvia chanryoenica is an endemic species, which locates on the ridges of mountains in South Korea (Chung et al. Citation2017). Although this plant has economic utility and importance as a genetic resource, molecular studies have not been conducted. Strategic research and conservation of important genetic resources are important. Chloroplast (cp) genomes are significantly resourced such as structural diversity (Sinn et al. Citation2018), molecular markers (Wang et al. Citation2018), and biogeographical inference (Ha et al. Citation2018). In this study, we reported the complete cp genome sequence of S. chanryoenica using next-generation sequencing technology. The plastid genome will contribute to develop protection strategy for this Endangered species.
S. chanryoenica was collected from Mt. Sobaek, South Korea. The voucher specimen was deposited at the herbarium of Korea National Arboretum (KH). Total genomic DNA was extracted using a DNeasy Plant MiniKit (Qiagen Inc., Valencia, CA, USA) and used for the next-sequencing with Illumina Miseq plat (Illumina, San Diego, California, USA). For the genome annotation, the online software DOGMA (Wyman et al. Citation2004) and tRNAscan-SE (Schattner et al. Citation2005) were used to adjust its difference by comparison with related genomes.
The cp genome of S. chanryoenica (Genbank accession number MH261357) is 151,689 bp in length and consists of a large (82,903 bp) and small (17,634 bp) single-copy regions, separated by a pair of identical inverted repeats (25,576 bp). This genome contains 79 unique protein-coding genes, 30 tRNA, and 4 rRNA. 18 genes encoded introns among unique genes of S. chanryoenica, among which 12 are protein-coding genes and six are tRNA genes. Three protein-coding genes include two introns (clpP, ycf3, and rps12), and the overall C + G content of S. chanryoenica is 37.9%. The gene order and organization of the S. chanryoenica are consistent with those of other Lamiaceae cp genomes.
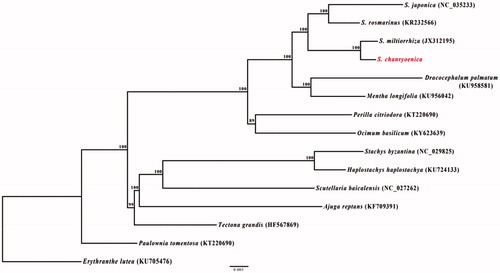
Phylogenetic analysis was conducted using a gene data matrix consisting of 71 protein-coding genes from 15 in Lamiaceae (). Erythranthe lutea and Paulownia tomentosa were used as an outgroup. Maximum likelihood tree was constructed with RAxML (Stamatakis Citation2014) using the GTR + R+I model with 1000 bootstrap replicates. The genus Salvia was well-supported monophyletic (100% bootstrap values, BS) and S. chanryoenica was sister to S. miltiorrhiza. Our results, the cp genome sequence of S. chanryoenica may contribute to a better understanding of the evolution of Salvia. Also offers a useful resource for molecular marker and species conservation.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chung GY, Chang KS, Chung J-M, Choi HJ, Paik W-K, Hyun J-O. 2017. A checklist of endemic plants on the Korean Peninsula. Korean J Pl Taxon. 47:264–288.
- Donoghue SM. 2002. Plant systematics: a phylogenetic approach. Sunderland (MA): Sinauer.
- Ha Y-H, Kim C, Choi K, Kim J-H. 2018. Molecular phylogeny and dating of Forsythieae (Oleaceae) provide insight into the Miocene history of Eurasian temperate shrubs. Front Plant Sci. 9:99.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Sinn BT, Sedmak DD, Kelly LM, Freudenstein JV. 2018. Total duplication of the small single copy region in the angiosperm plastome: rearrangement and inverted repeat instability in Asarum. Am J Bot. 105:71–84.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wang W, Chen S, Zhang X. 2018. Whole-genome comparison reveals heterogeneous divergence and mutation hotspots in chloroplast genome of Eucommia ulmoides Oliver. Int J Mol Sci. 19:1037.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhong G-X, Li P, Zeng L-J, Guan J, Li D-Q, Li S-P. 2009. Chemical characteristics of Salvia miltiorrhiza (Danshen) collected from different locations in China. J Agric Food Chem. 57:6879–6887.
- Zhou L, Zuo Z, Chow MSS. 2005. Danshen: an overview of its chemistry, pharmacology, pharmacokinetics, and clinical use. J Clin Pharmacol. 45:1345–1359.