Abstract
The complete mitchondrial genome of Echiniscus testudo, a cosmopolitan heterotardigrade collected in Japan, has been sequenced using a single molecule real-time (SMRT) sequencing long read after whole genome amplification from a single individual. The genome has a total length of 15,817 bp, consisting of 13 protein-coding genes, 20 tRNA, 2 rRNA genes, and an AT-rich control region. The nucleotide composition was extremely AT-rich, with 78.41% AT. Being in a different class of the phylum Tardigrada, there is a large sequence divergence from the previously reported mitochondrial genome of the eutardigrade Ramazzottius varieornatus, but the gene order of protein-coding genes are mostly conserved. This is the first report of a complete mitochondrial genome of a heterotardigrade.
Tardigrades are meiofaunal ecdysozoans with about 1200 species described to date. Of the two classes of tardigrades, heterotardigrades remains mostly unexplored through molecular studies, mostly due to the difficulty for sustainable culture in labs. Two genome sequences have been reported in eutardigrades (Arakawa et al. Citation2016; Hashimoto et al. Citation2016), but only an EST study have been reported thus far for heterotardigrades (Forster et al. Citation2009). Phylogenetic placement of Tardigrada within ecdysozoans still remains unsolved (Yoshida et al. Citation2017), and molecular studies of heterotardigrades would be a key data for this purpose. In this regard, here we present a complete mitochondrial genome sequence of a heterotardigrade Echiniscus testudo.
Single individual of E. testudo was isolated from a moss sample collected in Tsuruoka City, Japan (38.739641, 139.807600). The specimen is stored in the Institute for Advanced Biosciences, Keio University, Japan. After thoroughly washing the tardigrade to remove any remaining contaminants, high molecular weight DNA was extracted using MagAttract HMW DNA Kit (QIAGEN) and was amplified using Repli-G Midi Kit (QIAGEN). Purified DNA was then sequenced using PacBio RSII at Takara Bio. Longest read matching to mitochondrial genes was selected using BLAST searches (Altschul et al. Citation1997), and the read was subsequently error corrected with Illumina reads using Pilon (Walker et al. Citation2014). Circularity was checked manually, and the genome was annotated using MITOS2 WebServer (Bernt et al. Citation2013).
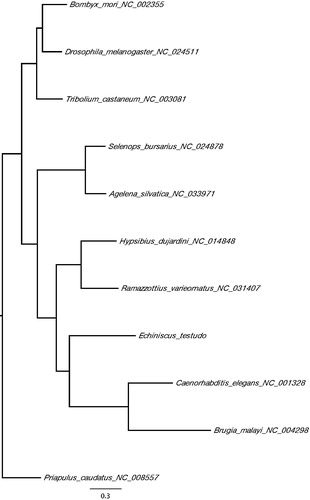
The complete mitochondrial genome sequence of E. testudo has a total length of 15,817 bp DDBJ accession number LC385650), consisting of 13 protein-coding genes, 22 tRNA, 2 rRNA genes, and an AT-rich control region. Because E. testudo belongs to a different class from previously sequenced tardigrade mitogenomes (Ramazzottius varieornatus and Hypsibius dujardini), sequence is highly diverged from these two genomes. On the other hand, gene order of protein-coding genes is surprisingly conserved. Phylogenetic analysis of the mitochondrial genomes of Tardigrada, Nematoda, and Arthropoda with Priapulida as an outgroup positions Tardigrada close to nematodes (). This is in line with previous phylogenetic studies using coding genes and provides a new perspective on the position of tardigrades within ecdysozoans.
Figure 1. A maximum-likelihood tree of the phylogenetic position of E. testudo among other ecdysozoan species. The tree was calculated from concatenated amino acid sequences of 13 mitochondrial protein genes using multiple alignment with MAFFT (Katoh and Standley Citation2014), followed by Trimal (80% consensus) and Fasttree (Price et al. Citation2009). Tree is visualized with FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Priapulus caudatus was used as an outgroup. GenBank accession numbers of mitogenome sequences used is shown after the species names.
Acknowledgments
The authors thank Nozomi Abe for technical assistance.
Disclosure statement
No potential conflict of interest was reported by the author.
Additional information
Funding
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402.
- Arakawa K, Yoshida Y, Tomita M. 2016. Genome sequencing of a single tardigrade Hypsibius dujardini individual. Sci Data. 3:160063.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Forster F, Liang C, Shkumatov A, Beisser D, Engelmann JC, Schnolzer M, Frohme M, Muller T, Schill RO, Dandekar T. 2009. Tardigrade workbench: comparing stress-related proteins, sequence-similar and functional protein clusters as well as RNA elements in tardigrades. BMC Genomics. 10:469.
- Hashimoto T, Horikawa DD, Saito Y, Kuwahara H, Kozuka-Hata H, Shin IT, Minakuchi Y, Ohishi K, Motoyama A, Aizu T, et al. 2016. Extremotolerant tardigrade genome and improved radiotolerance of human cultured cells by tardigrade-unique protein. Nat Comms. 7:12808.
- Katoh K, Standley DM. 2014. MAFFT: iterative refinement and additional methods. Methods Mol Biol. 1079:131–146.
- Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 26:1641–1650.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9:e112963.
- Yoshida Y, Koutsovoulos G, Laetsch DR, Stevens L, Kumar S, Horikawa DD, Ishino K, Komine S, Kunieda T, Tomita M, et al. 2017. Comparative genomics of the tardigrades Hypsibius dujardini and Ramazzottius varieornatus. PLoS Biol. 15:e2002266.
