Abstract
Penaeid shrimps are widely distributed from Indian to western Pacific Oceans and some which are economically important. In this study, we reported full mitochondrial genome of an endemic shrimp species, Penaeus acehensis, which inhabits exclusively in the coastal water of Aceh, Indonesia. Full length of circular mitogenome of P. acehensis was 15,991 bp in length, which contained 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and a control region. Start codons of all protein-coding genes were ATN except for COX1 in which ACG was used. Incomplete stop codon (T- -) was found in five genes including COX2, COX3, NAD5, NAD4, and NAD4L. Among its relatives, P. acehensis was most closely related to Penaeus monodon showing 89% sequence identity in its mitogenome, which was corresponding to morphological analysis. Phylogenetic tree result showed that P. acehensis was clustered together with those were distributed in Indo-West Pacific region (clade II), which is distinct from Eastern Pacific region (clade I).
Although penaeid shrimps are the economically important species in Southeast Asian countries, their native populations are being seriously threatened by the careless development in the coastal area (Páez-Osuna Citation2001). Penaeus acehensis was recently identified penaeid shrimp, which is exclusively distributed in Aceh province, Indonesia (Wedjatmiko Citation2009). Reddish body color without transverse band and unique numbers of rostral teeth (7–8) and ventral teeth (0–5) of P. acehensis are the morphological characteristics distinguished from its relatives including banana shrimp (Penaeus merguensis) or a black tiger shrimp (Penaeus monodon) (Alafanta Citation2014, Idami Citation2016, Wedjatmiko Citation2017).
In this research, the complete mitochondrial genome sequence of P. acehensis was determined using the combination of NGS and conventional PCR-based cloning methods. Voucher P. acehensis specimens were obtained from Brackishwater Aquaculture Development Center (BADC) in Ujung Batee, Aceh province, Indonesia. Genomic DNA was extracted from the muscle using an Accuprep Genomic DNA Extraction Kit (Bioneer) according to the manufacturer’s instruction. Full mitochondrial genome sequence of P. acehensis was obtained by assembling five fragmental PCR products generated by degenerated primer sets designed by the multiple alignments of mitogenome sequences from its relatives. For the sequencing, all PCR products were pooled together in equal concentration and fragmented into 350 bp in length by covaris M220 (Covaris Inc.). Thruseq® sample preparation kit version 2 (Illumina, USA) was used for the construction of a library and sequencing was performed using Illumina Miseq (Illumina, USA). Mothür software version 135.0 (Schloss et al. Citation2009) was used for pairing sequences and Geneious® 11.0.2 (Kearse et al. Citation2012) was used for mitogenome assembly.
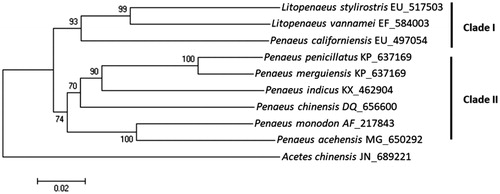
The total mitochondrial genome of P. acehensis is 15,991 bp in length (GenBank accession no. MG650292), which comprised 13 protein-coding genes, 22 transfer RNAs (RNAs), 2 ribosomal RNAs (rRNAs), and a putative control region. A + T content (71%) was higher than G + C content (29%) and the highest A + T content was observed in the putative control region (83%). Total 14 genes were located at L strand whereas the other remaining 23 genes were at H strands. Overlapping protein-coding genes were detected between ATP8 and ATP6 (7 bp), between ND4 and ND4L (19 bp). Each tRNA genes are predicted to be folded into a typical clover-leaf secondary structure except for tRNAser(GCT), which was predicted lacked D-arm. The phylogenetic trees were constructed based on minimum evolution algorithm, and Acetes chinensis was employed as an outgroup. Nucleotide sequence identity of P. acehensis ranged between 81% and 89%, and P. monodon was most closely related among the compared penaeid shrimps (). Phylogenetic analysis of penaeid shrimps showed two distinct clades (Clade I and II), which implicate a geographical distribution. Shrimps inhabit in Eastern Pacific waters including P. acehensis, P. monodon, and P. indicus were clustered into Clade I, whereas those inhabit in Indo west Pacific waters were clustered into Clade II.
Figure 1. Phylogenetic trees of Penaeus acehensis. The phylogenetic tree was constructed using molecular evolutionary genetic analyses (MEGA 6, version 6.0) with the minimum evolutionary algorithm. The evolutionary distance was calculated using Kimura 2 parameter method. Bootstrap replications were 1000. GenBank Accession number for each species was shown in bracket.
Acknowledgments
Authors thanks to Y. Heru Nugroho who helped with collecting the shrimp samples.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Alafanta D. 2014. Studi Fenotip Udang Pisang (Penaeus spp.) pada Hasil Budidaya. Aceh, Indonesia: Abulyatama University.
- Idami Z. 2016. Analisis Similaritas Karakter Morfologis dan Molekular Udang Pisang (Penaeus sp.) dengan Spesies Anggota Familia Penaeidae di Provinsi Aceh. Magister: Gadjah Mada University.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Páez-Osuna F. 2001. The environmental impact of shrimp aquaculture: causes, effects, and mitigating alternatives. Environ Manage. 28:131–140.
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. App Environ Microbiol. 75:7537–7541.
- Wedjatmiko W. 2017. Hasil tangkapan dan aspek biologi udang kelong (Penaeus sp.) di perairan barat Aceh. Jurnal Penelitian Perikanan Indonesia. 15:133–140.
