Abstract
The complete mitochondrial genome of Pacific abalone Haliotis discus hannai, an economically valuable shellfish in China, was sequenced and analyzed. The mitogenome was 16,716 bp in length and contained 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a putative control region. The gene arrangement and orientation were identical with other Haliotis species. Phylogenetic tree was constructed based on 13 PCGs of 7 Haliotidae species to assess their phylogenetic relationship, the tree revealed that H. discus hannai was more closely to H. rufescens and H. iris.
Pacific abalone Haliotis discus hannai is an economically important shellfish (Haliotidae, Gastropoda) in China, the production has reached 110,380 tons in 2013, which accounts for over 79% of the world abalone aquaculture output (Guo et al. Citation2017). In recent years, in order to improve yield, crossbreeding was used between Pacific abalone and other abalone (You et al. Citation2015), resulting in the contamination of Pacifc abalone germplasm. Thus, improving the molecular biology of Pacific abalone to facilitate germplasm management is extremely urgent.
The specimen of H. discus hannai was collected at Sanggou Bay (37°08′37.88″ N, 122°31′51.68″), China. After dissection, the foot muscle was deposited in the laboratory at −80 °C. The total DNA was extracted using a TIANamp Marine Animals DNA Kit (TIANGEN Biotech Co., Ltd.), and the whole mitochondrial genome was amplified with 24 pairs of primers. The mitochondrial sequence was assembled with DNAMAN software (Feng et al. Citation2014), and sequence annotation was performed using the online MITOS software (Bernt et al. Citation2013).
The complete mitogenome of Pacific abalone was 16,716 bp in length (GenBank Accession number: KU310896), with an AT bias of 60.4%, and contained 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a putative control region, shared 98.54% identities with Korea population (Yang et al. Citation2015). The gene arrangement and orientation are identical with other Haliotis species (Maynard et al. Citation2005; Xin et al. Citation2011; Robinson et al. Citation2016). Thirteen PCGs were 11,256 bp in length, encoding 3752 amino acids. All 13 genes were initiated by the start codon ATG, except for nad4 and nad5 (ATA), eight PCGs were terminated by a TAA codon, the genes in the nad4, nad4 L, cytb, nad6, and nad1 cluster were terminated by a TAG codon. The relative synonymous codon usage (RSCU) of Leu (15.6%) was the most frequent, while those encoding Cys (1.54%) and Arg (1.73%) were rare. The 22 tRNA genes vary from 63 to 72 bp in length. All of the tRNAs could be folded into typical cloverleaf secondary structures except tRNASer1, which lacked a DHU stem. A 7 bp amino acid acceptor stem was conserved in all tRNAs, except for tRNALys in which it was just 3 bp. The 16S rRNA and 12S rRNA were 1453 bp and 1009 bp, respectively, which were both shorter than that of Korea population (Yang et al. Citation2015).
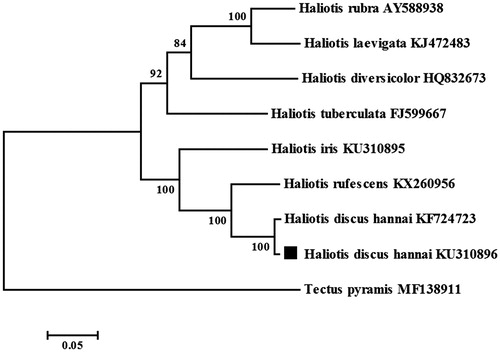
The phylogenetic analysis was performed using MEGA 6.0 software (Tamura et al. Citation2013) with 1000 bootstrap replicates based on the concatenated nucleotide sequences of the 13 PCGs of 7 Haliotis species (including H. discus hannai Korea population) available in GenBank, Tectus pyramis was used as an outgroup (). The phylogenetic tree revealed that 7 Haliotis species constituted two major subclades, H. discus hannai (Chinese population and Korea population), H. rufescens, and H. iris clustered in one clade, the rest of Haliotis species formed the second clade, which was similar to the result based on ITS gene (Guo et al. Citation2017).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Feng S, Powell SM, Wilson R, Bowman JP. 2014. Extensive gene acquisition in the extremely psychrophilic bacterial species Psychroflexus torquis and the link to sea-ice ecosystem specialism. Genome Biol Evol. 6:133–148.
- Guo ZS, Ding Y, Zhang XH, Hou XG. 2017. Complete nuclear ribosomal DNA sequence analysis of Pacifc abalone Haliotis discus hannai. Fish Sci. 83:777–784.
- Maynard BT, Kerr LJ, McKiernan JM, Jansen ES, Hanna PJ. 2005. Mitochondrial DNA sequence and gene organization in the [corrected] Australian blacklip [corrected] abalone Haliotis rubra (leach). Mar Biotechnol. 7:645–658.
- Robinson NA, Hall NE, Ross EM, Cooke IR, Shiel BP, Robinson AJ, Strugnell JM. 2016. The complete mitochondrial genome of Haliotis laevigata (Gastropoda: Haliotidae) using MiSeq and HiSeq sequencing. Mitochondrial DNA A DNA Mapp Seq Anal. 27:437–438.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Xin Y, Ren JF, Liu X. 2011. Mitogenome of the small abalone Haliotis diversicolor Reeve and phylogenetic analysis within Gastropoda. Mar Genom. 4:253–262.
- Yang EC, Nam BH, Noh SJ, Kim YO, Kim DG, Jee YJ, Park JH, Noh JH, Yoon HS. 2015. Complete mitochondrial genome of Pacific abalone (Haliotis discus hannai) from Korea. Mitochondrial DNA. 26:917–918.
- You WW, Guo Q, Fan FL, Ren P, Luo X, Ke CH. 2015. Experimental hybridization and genetic identification of Pacific abalone Haliotis discus hannai and green abalone H. fulgens. Aquaculture. 448:243–249.