Abstract
In this study, the complete mitochondrial genome of Garrulax milnei was sequenced for the first time. This mitogenome is 17,871bp in length and it contains 22 tRNA genes, 13 protein-coding genes, 2 rRNA genes, 1 control region and 1 extra pseudo-control region. The base composition of the mitochondrial genome is 29.7% A, 24.5% T, 14.2% G and 31.6% C. The phylogenetic analysis based on the 12 protein-coding genes (except for ND6 gene) showed that G. milnei clusters with other two species that belong to genus Garrulax.
The Garrulax milnei, also called Trochalopteron milnei, is classified under the order Passeriformes, family Leiotrichidae (BirdLife International Citation2016). In this study, the muscle sample of G. milnei was collected through the field investigation in Guangzhou province, China, and the geo-spatial coordinates are 23°09′ 52″N latitude, and 1133°30′ 40″E longitude. The samples are deposited in the Herbarium of the Institute of Protection and Utilization for Biological Resource at Qufu Normal University in Shandong province, China. The sample died naturally and DNA was extracted with the DNeasy Blood & Tissue kit (QIAGEN, U.S.A) according to the manufacturer’s protocol. After assembled and annotated, this sequence has been deposited in GenBank with the accession No. MH238447.
The complete mitochondrial genome of G. milnei is 17,871 bp in length and encodes 37 genes which contains 22 transfer RNA genes (tRNA), 13 protein-coding genes (PCGs), and 2 ribosomal RNA genes (rRNA). Among these genes, 9 genes (ND6,tRNAAla, tRNAAsn, tRNACys, tRNAGln, tRNAGlu, tRNAPro, tRNASer, tRNATyr) encoded in L-strand, and other genes encoded in H-strand. The base composition is 29.7% for A, 24.5% for T, 14.2% for G, 31.6% for C and the percentage of A and T (54.2%) is higher than that of G and C (45.8%). The gene structure, content, and arrangement are similar to other Garrulax sequenced in the early research (Huang and Zeng Citation2016).
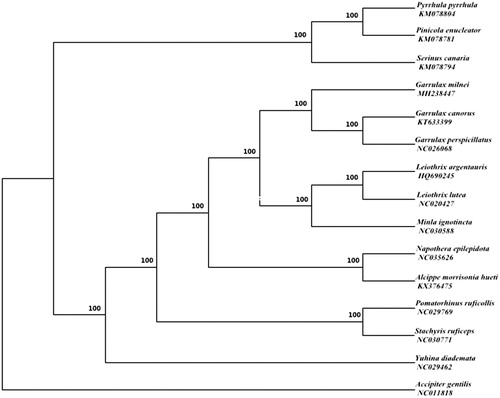
Phylogenetic relationships of G. milnei and 13 other species were analyzed with Bayesian inference (BI) methods based on 12 protein-coding genes except for ND6, and the Accipiter gentilis (NC011818) was chosen as an out-group. According to the AIC test criterion, GTR + I + G was selected as the optimal evolutionary model (Tracey et al. Citation2007) by MrBayes 3.1.2 (Ronquist and Huelsenbeck Citation2003) and the posterior probabilities are shown on the nodes of the tree. The result of phylogenetic tree showed that G. milnei clusters with other two species that belong to Garrulax (Figure 1). To date, few studies have been recorded for Garrulax, and we hope that our study can provide a useful database for further research.
Figure 1. Bayesian phylogenetic inference (BI) trees of 14 species based on 12 protein-coding genes except for ND6 and the posterior probabilities are shown on the nodes. The accession numbers of 14 species are KM078804 (P.pyrrhula), KM078781 (P.enucleator), KM078794 (S.canaria), KT633399 (G.canorus), NC026068 (G.perspicillatus), HQ690245 (L. argentauris), NC020427 (L.lutea), NC030588 (M.ignotincta), NC035626 (N.epilepidota), KX376475 (A.M.hueti), NC029769 (P.ruficollis), NC030771 (S.ruficeps), NC029462 (Y.diademata), NC011818 (A.gentilis).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- BirdLife International. 2016. Trochalopteron milnei. The IUCN Red List of Threatened Species 2016: e.T22715764A94468047. Downloaded on 24 April 2018. http://www.birdlife.org
- Huang L, Zeng B. 2016. Complete mitochondrial genome of the Chinese hwamei (Garrulax canorus). Mitochondrial DNA. 27:4559–4560.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Tracey SR, Smolenski A, Lyle JM. 2007. Genetic structuring of Latris lineataat localized and transoceanic scales. Mar Biol. 152:119–128.
