Abstract
The two-spot swimming crab Charybdis bimaculata (Miers, Citation1886) is an important decapod species in the benthic ecosystem of Korean waters. In this study, we determined its complete mitochondrial genome by the combination of NGS analysis using MiSeq platform and PCR-based cloning method. The circular mitochondrial genome of C. bimaculata was 15,714 bp in length in which the standard set of 13 protein-coding genes, 22 tRNA genes, and 2 rRNA genes were encoded. Phylogenic analysis showed that C.bimaculata is most closely related to Charybdis feriata. The complete mitogenome sequence information of C. bimaculata would provide useful data for the conservation of their population in the Pacific ocean.
Charybdis bimaculata (Miers, Citation1886) is a small portunid crab, which is mainly caught in Korea, China, and Japan (Rho et al. Citation2005; Gomez et al. Citation2008; Narita et al. Citation2008; Kwak et al. Citation2014). Although it is widely distributed and plays an important role in the benthic ecosystem (Kume et al. Citation1999; Yamaguchi and Taniuchi Citation2000), ecological study of C. bimaculata has been limited mainly due to its lack of genetic information. We here determined the complete mitochondrial genome of C. bimaculata using next generation sequencing (NGS) platform.
The specimen was collected from Jindo Island (34°22′42.62″N and 126°8′14.71″E), morphological identification was determined by National Institute of Fisheries Science, Korea. The specimen or its DNA was stored at Pukyong National University. Two mitochondrial DNA fragments were amplified by two degenerated primer sets targeting COI-ND5 and ND5-COI regions, respectively, and D-loop region was amplified by a sequence-specific primer set. Three PCR products were then pooled together in equal concentration and fragmented into smaller fragments (∼350 bp) by Covaris® M220 Focused-UltrasonicatorTM (Covaris Inc., U.S.A). Library was constructed by TruSeq® RNA library preparation kit V2 (Illumina, USA). After confirming the quality and the quantity using 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, U.S.A), DNA sequencing was performed by MiSeq sequencer (2 × 300 bp ends) (Illumina, U.S.A).
The complete mitochondrial genome of C. bimaculata (GenBank Number: MG489891) was 15,714 bp which encodes 13 proteins, 22 tRNAs, 2 rRNAs and a putative control region (D-loop). Among them, 12 genes were initiated by the start codon ATN (ATG and ATT) with an exception of ATP8 gene, which begins with GTG. The canonical stop codons (TAA or TAG) were shown in nine genes (ATP8, ATP6, ND2, ND3, ND4, ND4L, ND5, and ND6), whereas incomplete stop codons (T–– or TA–) were found in the other four (COI, COII, COIII, and Cyt b) genes. Twenty one tRNA were predicted to be folded into a typical clover-leaf secondary structures by ARWEN (Laslett and Canbäck 2007) except for tRNASer, which was predicted without DHC arm.
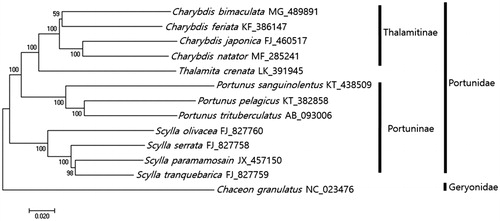
The phylogenetic position of C. bimaculata within portunid crabs was analyzed using minimum evolutionary (ME) methods (). As the result of phylogenetic tree which was constructed with the full mitochondrial genome sequences from 12 portunids and one Geryonid as out group, all four Charybdis species including C. bimaculata, C. feriata (Ma et al. Citation2015), C. natator (Yang et al. Citation2017), and C. japonica (Liu and Cui Citation2010) were clustered together. C. bimaculata was most closely related to C. feriata showing 86% sequence identity (). This information would be useful for extending our knowledge about evolutional relationship of Portunidae species.
Figure 1. Phylogenetic tree of Charybdis bimaculata with other Portunid crabs. Phlyogenetic tree of Charybdis bimaculata was constructed with mitochondrial genomes of its relative species using MEGA7 software with Minimum Evolution (ME) algorithm with 1000 boothstrap replications. Chaceon granulatus in Geryonidae was adopted as the out group member. GenBank Accession numbers were shown followed by each scientific name.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Gomez DK, Baeck GW, Kim JH, Choresca CH, Park SC. 2008. Molecular detection of betanodaviruses from apparently healthy wild marine invertebrates. J Invertebr Pathol. 97:197–202.
- Kume G, Yamaguchi A, Taniuchi T. 1999. Feeding habits of the cardinal fish Apogon lineatus in Tokyo Bay, Japan. Fish Sci. 65:420–423.
- Kwak S-N, Park J-M, Huh S-H. 2014. Seasonal variations in species composition and abundance of fish and decapods in an eelgrass (Zostera marina) bed of Jindong Bay. J Korean Soc Mar Environ Saf. 20:259–269.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Liu Y, Cui Z. 2010. Complete mitochondrial genome of the Asian paddle crab Charybdis japonica (Crustacea: Decapoda: Portunidae): gene rearrangement of the marine brachyurans and phylogenetic considerations of the decapods. Mol Bio Rep. 37:2559–2569.
- Ma H, Ma C, Li C, Lu J, Zou X, Gong Y, Wang W, Chen W, Ma L, Xia L. 2015. First mitochondrial genome for the red crab (Charybdis feriata) with implication of phylogenomics and population genetics. Sci Rep. 5:
- Miers EJ. 1886. Report on the Brachyura collected by HMS Challenger during the years 1873-76. Rep scient Results Voy Challenger (Zool). 17:1–29.
- Narita T, Ganmanee M, Sekiguchi H. 2008. Population dynamics of portunid crab Charybdis bimaculata in Ise Bay, central Japan. Fish Sci. 74:28–40.
- Rho HS, Jung J, Song SJ, Kim W. 2005. Crustacean decapods of Jindo Island, Korea. Anim Syst Evol Divers. 5:1–14.
- Yamaguchi A, Taniuchi T. 2000. Food variations and ontogenetic dietary shift of the starspotted‐dogfish Mustelus manazo at five locations in Japan and Taiwan. Fish Sci. 66:1039–1048.
- Yang X, Ma H, Waiho K, Fazhan H, Wang S, Wu Q, Shi X, You C, Lu J. 2017. The complete mitochondrial genome of the swimming crab Charybdis natator (Herbst) (Decapoda: Brachyura: Portunidae) and its phylogeny. Mitochondrial DNA Part B. 2:530–531.
