Abstract
Rumex acetosa, known as sheep’s sorrel, red sorrel, sour weed, and field sorrel, is a species of flowering plant in the buckwheat family Polygonaceae. In this study, the complete chloroplast (cp) genome of R. acetosa (Rumiceae) was determined through Illumina sequencing method. The complete chloroplast genome of R. acetosa was 160,269 bp in length and contained a pair of IR regions (30,503 bp) separated by a small single copy region (13,128 bp) and a large single copy region (86,135 bp). This cp genome is encoded with 129 genes including 83 protein-coding genes, 36 tRNA genes, and 8 ribosomal RNA genes. The overall GC content of R. acetosa cp genome is 37.2%. By phylogenetic analysis using Bayesian method, R. acetosa showed the closest relationship with other 2 Rumiceae species, Rheum palmatum and Oxyria sinensis.
Rumex acetosa L. (Polygonaceae), a dioecious plant which has a multiple sex chromosome system (Shibata et al. Citation1999, Citation2000), is a perennial herb, commonly known as sheep’s sorrel, red sorrel, sour weed, and field sorrel (Lee et al. Citation2005). As a traditional medicine plant, it was shown to have some pharmacological activities, including anti-inflammatory, antioxidant (Wegiera et al. Citation2007), anti-tumor, antibacterial, antiviral, and anti-fungal properties (Taylor et al. Citation1996; Demirezer et al. Citation2001; Lee et al. Citation2005). Meanwhile, its leaves are widely used in sauces and salads (Ahmad et al. 2006). With an aim to retrieve valuable cp molecular markers, indels, and SSRs by comparative analyses with other Rumiceae cp genomes, we assembled and analyzed the chloroplast genome of R. acetosa based on the next-generation sequencing method.
Leaves from R. acetosa were collected in Sierra Nevada, Granada, Spain (37°10′N, 3°17′W). Both extracted DNA and this species voucher were stored at Guangxi Botanical Garden of Medicinal Plants. Sequencing was done on the Illumina Hiseq-2500 platform to produce 150 bp paired-end reads (BGI Tech, Shenzhen, China). After reads quality filtration, the clean reads were assembled by SPAdes 3.6.1 (Bankevich et al. Citation2012). We used chloroplast genome of Rheum palmatum (accession number: KR816224) (Fan et al. Citation2015) as a reference sequence to align the contigs and identify gaps. To fill the gap, Price (Ruby et al. Citation2013) and MITObim v1.8 (Hahn et al. Citation2013) were applied, and Bandage (Wick et al. Citation2015) was used to identify the borders of the IR, LSC, and SSC regions. The complete sequence was primarily annotated by Geseq (Tillich et al. Citation2017) and Plann (Huang et al. Citation2015) combined with manual correction. All tRNAs were confirmed using the tRNAscan-SE search server (Lowe et al. Citation1997). Protein-coding genes were verified by BLAST search on the NCBI website (http://blast.ncbi.nlm.nih.gov/) and manual correction for start and stop codons was conducted. The circular cp genome map was drawn using OrganellarGenomeDRAW (Lohse et al. Citation2007). This complete chloroplast genome sequence together with gene annotations were submitted to GenBank under the accession number of MH359405.
The chloroplast genome of R. acetosa is a typical quadripartite structure with a length of 160,269 bp. The whole cp genome contains a large single-copy (LSC) region of 86,135 bp, a small single-copy (SSC) region of 13,128 bp, and 2 inverted repeat (IRs) regions of 30,503 bp. The cp genome possesses 129 genes, including 83 protein-coding genes (78 PCG species), 8 ribosomal RNA genes (4 rRNA species) and 36 tRNA genes (30 tRNA species). The overall GC content of the cp genome is 37.2%. The genome structure, gene order, and GC content are similar to other Rumiceae cp genomes.
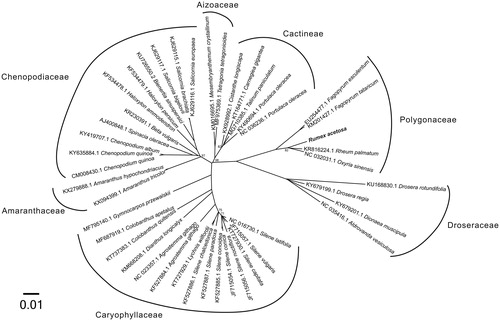
For phylogenetic analysis assessing the relationship of this plastid, we selected other 43 Caryophyllales cp genomes including Caryophyllaceae (15 taxa), Amaranthaceae (2 taxa), Chenopodiaceae (11 taxa), Aizoaceae (2 taxa), Cactineae (5 taxa), Polygonaceae (4 taxa), and Droseraceae (4 taxa) to construct a genome-wide alignment. We considered plastids of the Fagales as the outgroup. The genome-wide alignment of all cp genomes was done by HomBlocks (Bi et al. Citation2018), resulting in a total of 45,627 positions. The whole genome alignment was analyzed by PhyloBayes ver. 3.3 (Lartillot et al. Citation2009) under the CAT-GTR + Γ model that accounts for across-site heterogeneities. Four independent MCMC analyses were run for 1,000,000 cycles in PhyloBayes. Convergence was verified based on time-series plots of the likelihood scores using Tracer (http://tree.bio.ed.ac.uk/software/tracer/). The first 25% cycles were discarded as burn-in, and the maximum clade credibility (MCC) tree was constructed in TreeAnnotator v1.8.0 (Rambaut et al. 2012) depicting the maximum sum of Bayesian posterior probabilities (BPPs). The resulting tree was represented and edited using FigTree v1.4.1 (http://tree.bio.ed.ac.uk/software/fgtree/). As shown in , the phylogenetic positions of these 44 cp genomes were successfully resolved with full BPPs supports except for 6 nodes. Rumex acetosa belongs to the Polygonaceae as expected and exhibited the closest relationship with other 2 Rumiceae species, Rheum palmatum and Oxyria sinensis.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Ahmad S, Ullah F, Sadiq A, Ayaz M, Imran M, Ali I, Zeb A, Ullah F, Shah MR. 2016. Chemical composition, antioxidant and anticholinesterase potentials of essential oil of Rumex hastatus D. Don collected from the North West of Pakistan. BMC Complementary Altern Med. 16(1):29.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. JComp Bio. 19:455–477.
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22.
- Demirezer LÖ. 2001. The structures of antioxidant and cytotoxic agents from natural source: anthraquinones and tannins from roots of Rumex patientia. Phytochemistry. 58:1213–1217.
- Fan K, Sun X-J, Huang M, Wang X-M. 2015. The complete chloroplast genome sequence of the medicinal plant Rheum palmatum L.(Polygonaceae). Mitochondrial DNA Part A. 27:1–2936.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Lartillot N, Lepage T, Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 25:2286–2288.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 41(13):e129–e129.
- Lee N-J, Choi J-H, Koo B-S, Ryu S-Y, Han Y-H, Lee S-I, Lee D-U. 2005. Antimutagenicity and cytotoxicity of the constituents from the aerial parts of Rumex acetosa. Biol Pharm Bull. 28:2158–2161.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Rambaut A, AJ. Drummond TreeAnnotator version 1.6. 1. University of Edinburgh, Edinburgh, UK. Available at: http://beast. bio. ed. ac. uk (2010).
- Ruby JG, Bellare P, DeRisi JL. 2013. PRICE: software for the targeted assembly of components of (Meta) genomic sequence data. G3: Genes| Genomes| Genetics. 3:865–880.
- Shibata F, Hizume M, Kuroki Y. 1999. Chromosome painting of Y chromosomes and isolation of a Y chromosome-specific repetitive sequence in the dioecious plant Rumex acetosa. Chromosoma. 108:266–270.
- Shibata F, Hizume M, Kuroki Y. 2000. Differentiation and the polymorphic nature of the Y chromosomes revealed by repetitive sequences in the dioecious plant, Rumex acetosa. Chromosome Res. 8:229–236.
- Taylor RSL. 1996. Antiviral activities of medicinal plants of southern Nepal. J Ethnopharmacol. 53:97–110.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes Nucleic Acids Res. 31(20):3350–3352.
- Wegiera M, Smolarz HD, Wianowska D, Dawidowicz AL. 2007. Anthracene derivatives in some species of Rumex L. genus. Acta Soc Bot Pol. 76:103.
- Wick RR, et al. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics.