Abstract
The complete mitochondrial genome was sequenced in two individuals of the Arctic rainbow smelt Osmerus dentex. The genome sequences are 16,615 and 16,616 bp in size, and the gene arrangement, composition, and size are very similar to the other smelt mitochondrial genomes published previously. The difference between the two O. dentex genomes studied is 0.25%, that is, 5.0 times higher in comparison with close species, the European smelt O. eperlanus studied previously. The level of mitochondrial genome divergence between O. dentex and close osmerid fishes, O. eperlanus, and O. mordax is high enough (6.86–7.54%) to consider all of them as separate biological species.
The Arctic rainbow smelt Osmerus dentex Steindachner and Kner has wide distribution in the North Pacific regions from the Wonsan coastal area (North Korea) and the Sea of Okhotsk to Barkley Sound (British Columbia), northward to the Bering Sea, and the Arctic Ocean from the White Sea to the Chukotka Sea (eastern Siberia) (Kottelat and Freyhof Citation2007). Berg (Citation1949) considered O. dentex as a subspecies of the European smelt O. eperlanus (L.). McAllister (Citation1963) distinguished two subspecies, O. eperlanus eperlanus and O. eperlanus mordax sensu lato including O. eperlanus dentex. Kljukanov and McAllister (Citation1973) concluded that O. mordax consists of two subspecies, O. mordax mordax and O. mordax dentex (https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=8013).
Using the mitochondrial (mt) DNA restriction fragment length and cytochrome b sequence polymorphism, Taylor and Dodson (Citation1994) revealed a high level of divergence (5.6–8.9%) between the Arctic, European, and Atlantic smelts suggesting the species status for each of them. The morphological data (Shedko Citation2001) presented new arguments to increase the O. dentex status from subspecies to species level. However, the scientific literature and particularly the GenBank Taxonomy Browser still list O. dentex as a subspecies of O. mordax.
To increase the power of the molecular taxonomy analysis of this complex fish group we have sequenced two complete mt genomes of O. dendex (GenBank accession numbers MH370836 and MH370837) from the Amur Bay of the Sea of Japan (43˚13′ 17,256″ N; 131˚55′ 37,113″ E). The primers were designed with the program mitoPrimer_V1 (Yang et al. Citation2011). The fish specimens are stored at the museum of the National Scientific Center of Marine Biology, Vladivostok, Russia (www.museumimb.ru) under accession numbers MIMB35006 and MIMB35007.
The O. dentex mt genome sequences are 16,615 and 16,616 bp in size and the gene arrangement, composition, and size are very similar to the smelt fish genomes published previously. We detected 41 single nucleotide and one length differences between the haplotypes 394Omd1 and 397Omd4; total sequence divergence (Dxy) was 0.0025 ± 0.0004. The difference between the two O. dentex genomes studied is relatively high in comparison with close species, the European smelt O. eperlanus, which demonstrated 5.0 times smaller difference (0.0005 ± 0.0001) between two genomes studied previously (Balakirev et al. Citation2018), which corresponds with much wider species range in O. dentex than in O. eperlanus (McAllister Citation1984; Kottelat and Freyhof Citation2007), but it could also indicate the overfishing in O. eperlanus populations (Hutchinson and Mills Citation1987).
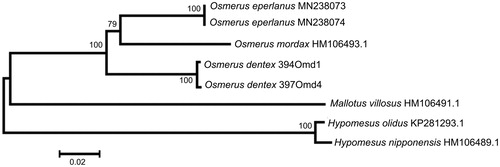
The comparison of mt genomes now obtained with other complete mt genomes of related groups available in GenBank including genera Hypomesus, Osmerus, and Mallotus reveals a close affinity of O. dentex to other Osmerus species (). The level of sequence divergence between O. dentex, O. eperlanus, and O. mordax is still high enough (6.86–7.54%) to consider all of them as separate biological species. Similar values of interspecific divergence between osmerid fishes were reported previously (e.g. Skurikhina et al. Citation2013 and references therein).
Figure 1. Maximum likelihood tree for the Arctic rainbow smelt Osmerus dentex specimens 394Omd1 and 397Omd4, and GenBank representatives of the order Osmeriformes. The tree is based on the General Time Reversible + gamma + invariant sites (GTR + G + I) model of nucleotide substitution. The numbers at the nodes are bootstrap percent probability values based on 1000 replications.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Balakirev ES, Kravchenko AYu, Romanov NS, Ayala FJ. 2018. Complete mitochondrial genome of the European smelt Osmerus eperlanus (Osmeriformes, Osmeridae). Mitochondrial DNA B.
- Berg LS. 1949. Freshwater fish of the USSR and adjacent countries. Moscow: Akad. Nauk SSSR.
- Hutchinson P, Mills DH. 1987. Characteristics of spawning-run smelt, Osmerus eperlanus (L.), from a Scottish river, with recommendations for their conservation and management. Aquaculture Res. 3:744–755.
- Kljukanov VA, McAllister DE. 1973. Osmeridae. In: Hureau J-C, Monod T, editors. Check-list of the fishes of the north-eastern Atlantic and of the Mediterranean (CLOFNAM). Vol. 1. Paris: UNESCO. p.158–159.
- Kottelat M, Freyhof J. 2007. Handbook of European freshwater fishes. Berlin: Publications Kottelat, Cornol and Freyhof.
- McAllister DE. 1963. A revision of the smelt family, Osmeridae. Bull Nat Mus Can. 191:1–53.
- McAllister DE. 1984. Osmeridae. In: Whitehead PJP, Bauchot M-L, Hureau J-C, Nielsen J, Tortonese E, editors. Fishes of the north-eastern Atlantic and the Mediterranean. Paris: UNESCO. Vol. 1. p. 399–402.
- Shedko SV. 2001. On species composition of smelts of Maritime territory. Voprosy Ichtiologii. 41:261–264.
- Skurikhina LA, Kukhlevsky AD, Kovpak NE. 2013. Relationships of osmerid fishes (Osmeridae) of Russia: divergence of nucleotide sequences of mitochondrial and nuclear genes. Genes Genom. 35:529–539.
- Taylor EB, Dodson JJ. 1994. A molecular analysis of relationships and biogeography within a species complex of Holarctic fish (genus Osmerus). Mol Ecol. 3:235–248.
- Yang CH, Chang HW, Ho CH, Chou YC, Chuang LY. 2011. Conserved PCR primer set designing for closely-related species to complete mitochondrial genome sequencing using a sliding window-based PSO algorithm. PLoS One. 6:e17729
