Abstract
Paeonia lactiflora has been listed as an Endangered species in Russian Federation. The complete plastome was assembled from Next-Generation Sequencing data. It is 152,747 bp in length. It consists of a pair of Inverted Repeat regions (25,651 bp), separated by a small single copy region of 17,033 bp and a large single copy region of 84,412 bp. The plastome encoded 128 genes, including 83 protein coding genes, 37 tRNA, eight rRNA genes, four pseudogenes, and is characterized by loss of the rpl32 and infA genes. Phylogenetic analysis of Paeoniaceae plastomes revealed that P. lactiflora clustered with Eurasian peonies (section Paeoniae).
Paeonia lactiflora (Pallas Citation1776) is an important ornamental plant; it also has a long history of use as traditional medicinal and oil plant (Tsai et al. Citation2008; Zhou et al. Citation2011). This species grows in meadow slopes and foothills in China, Korea, Mongolia, and the Russian Federation (Far East, Siberia). P. lactiflora has been assessed as Endangered to the Russian Far East (Red data book Primorsky Krai Citation2008; The Red Book of the Russian Federation (plants and fungi) Citation2008). Climate fluctuation and the reduction of occupied area has resulted in the increasingly declining wild populations of the P. lactiflora mature individuals. The comprehensive genomic information is very important to formulate effective conservation strategies for this valuable wild peony species. Here, the complete plastome of P. lactiflora was determined using next-generation sequencing technology. The assembled and annotated plastome was deposited in GenBank under the accession number MG897127.
Paeonia lactiflora plants were collected in Primorsky Krai, Russia (43 16.512' N, 134 03.144' E.). A voucher specimen (41-3 MW) was deposited in the herbarium of MSU, Russian Federation.
Total DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle Citation1987). The total DNA (1 μg) was used to build the paired-end libraries. The library was sequenced using an Illumina HiSeq 2000 instrument (Illumina, Inc., USA). The resulting readings were processed using the CLC Genomics Workbench software v. 5.5. (http://www.clcbio.com). The assembled plastome sequence was annotated using the online tool CpGAVAS (Liu et al. Citation2012) with default options and then checked manually.
The plastome of P. lactiflora is 152,747 bp in length consisting of a pair of inverted repeat regions (25,651 bp) separated by a small single copy region (17,033 bp) and a large single copy region (84,412 bp). The plastome encoded 128 genes, including 83 protein-coding genes, 37 tRNA, eight rRNA genes, and four pseudogenes: rps19, ycf1, rps18, rpl22 and is characterized by loss of the rpl32 and infA genes. The overall GC content is 38.4%.
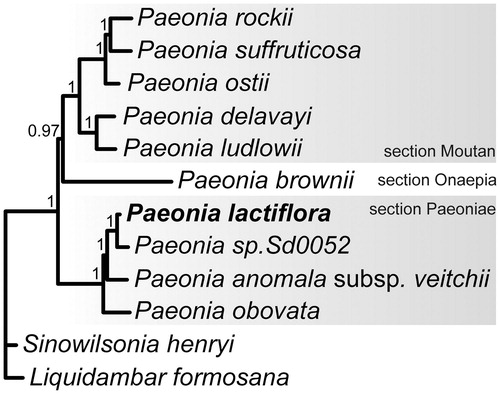
The phylogenetic tree was reconstructed using 10 published complete Paeoniaceae and two Saxifragales plastomes as the outgroup. The IRa region was removed from analysis, sequences were aligned using MUSCLE (Edgar Citation2004), and variable and gap-rich positions were removed from alignment using GBLOCKS (Castresana Citation2000). Phylogeny inference was performed applying the Bayesian approach using MrBayes (Ronquist et al. Citation2012). Phylogenetic analysis produced a fully resolved tree with P. lactiflora clustered with Eurasian peonies (section Paeoniae) ().
Figure 1. Bayesian tree based on 12 complete plastome sequences. Accession numbers: P. lactiflora (MG897127, this study), Paeonia sp. Sd0052 (KF753636), P. anomala subsp. veitchii (NC_032401), P. obovata (NC_026076), P. ludlowii (NC_035623), P. delavayi (NC_035718), P. ostii (NC_036834), P. suffruticosa (MH_191384), P. rockii (MF488719), P. brownii (MH191385), Sinowilsonia henryi (NC_036069), and Liquidambar formosana (NC_023092).
Acknowledgements
The authors thank Richard H. Lozier for editing this manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Pallas PS. 1776. Reise durch Verschiedene Provinzen des Russischen Reichs. St. Petersburg: Kaiserliche Academie der Wissenschaften
- Red Data Book Primorsky Krai. 2008. Plants rare and Endangered species of plants and fungi. Official edition. Vladivostok. p. 688 (in Russian).
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- The Red Book of the Russian Federation (plants and fungi). 2008. The Ministry of Natural Resources and Environment of Russian Federation. Moscow: KMK Scientific Press Ltd. p. 885 (in Russian).
- Tsai H-Y, Lin H-Y, Fong Y-C, Wu J-B, Chen Y-F, Tsuzuki M, Tang Ch H. 2008. Paeonol inhibits RANKL-induced osteoclastogenesis by inhibiting ERK, p38 and NF-kappaB pathway. Eur J Pharmacol. 588:124–133.
- Zhou C, Zhang Y, Sheng Y, Zhao D, Lv S, Hu Tao J. 2011. Herbaceous peony (Paeonia lactiflora Pall.) as an alternative source of oleanolic and ursolic acids. Int J Mol Sci. 12:655–667.
