Abstract
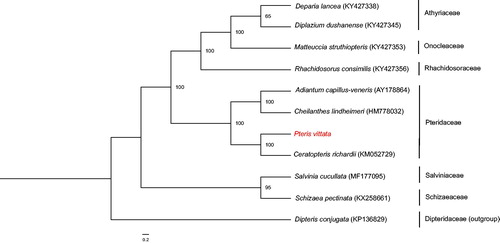
It is the first report on complete chloroplast genome of Pteris vittata, an arsenic hyperaccumulating fern. Its genome size is 154,130 bp, with a typical circular structure including a large single-copy (LSC) (82,623 bp) and a small single-copy (SSC) (20,957 bp) regions separated by a pair of inverted repeats (25,275 bp each). The plastome encodes 132 genes, including 87 protein-coding genes, 35 tRNA genes, eight rRNA genes, and two pseudogenes. The overall Guanine+Cytosine (GC) content is 41.7% and GC content in the IR regions is higher than in the LSC and SSC regions. Maximum likelihood (ML) tree indicated that P. vittata was clustered with Ceratopteris richardii.
Pteris vittata, commonly known as Chinese brake or ladder fern, is a terrestrial fern belonging to Pteridaceae. It has erect rhizomes and dark straw-coloured fronds. As a perennial evergreen species native to China, it is widely distributed in tropics and subtropics of the Old World (Zhang et al. Citation2013). The fern grows in calcareous soils, on limestone, also on concrete structures and cracks at an altitude of below 2000 m (Zhang et al. Citation2013). It is not only a traditional Chinese medicine used for influenza and dysentery (Xie Citation1996) but also a famous hyperaccumulator of arsenic in phytoremediation (Cesaro et al. Citation2015). In addition, relationships among some groups in Pteridaceae are subjected to be further classified (Christenhusz and Chase Citation2014; Ruhfel et al. Citation2018). Therefore, acquirement of the complete chloroplast of P. vittata has important implication for investigating role of chloroplast in arsenic accumulation and deeper phylogenetic relationships.
We isolated total genomic DNA from the fresh leaves of P. vittata collected from South China Botanical Garden, Chinese Academy of Sciences (23°11′3.56″N, 113°21′43.28″E), using Tiangen Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China) according to the instructions of the manufacturer. Voucher specimen was deposited in the Herbarium of Sun Yat-sen University (SYS; voucher: SS Liu 201616). After an average of 300 bp, genomic DNA library was constructed, pair-end sequencing was generated on an Illumina Hiseq 2500 platform. We obtained 7,077,109 raw reads. After trimming the sequences, 5,494,147 clean reads were de novo assembled into complete chloroplast genome by Velvet v1.2.07 (Zerbino and Birney Citation2008). The protein-coding genes, tRNA genes, and rRNA genes were predicted using DOGMA (Wyman et al. Citation2004) and tRNAscan-SE (Schattner et al. Citation2005). Maximum likelihood (ML) analysis was performed through RAxML v8.0 (Stamatakis Citation2014) with 1000 bootstrap replicates using 11 ferns including Dipteris conjugate as an outgroup aligned with MAFFT v.7.221 (Katoh and Standley Citation2013).
The whole chloroplast genome of P. vittata (GenBank Accession Number: MH500228) has a total length of 154,130 bp, with 41.7% GC content. It is a typical quadripartite structure with a large single-copy (LSC) region of 82,623 bp, a small single-copy (SSC) region of 20,957 bp, and a pair of inverted repeats (IRa and IRb) of 25,275 bp. The plastome encodes 132 genes, including 87 protein-coding genes, 35 tRNA genes, eight rRNA genes, and two pseudogenes (ndhB and cemA). The IR regions have higher GC content (45.0%) than the LSC (40.6%) and SSC (37.7%) regions. Most genes occur as a single copy, except for four protein-coding genes, six tRNA genes, and four rRNA genes, which are duplicated in the IR regions. In addition, only three genes (ycf3, clpP, and rps12) have two introns. ML tree indicated that P. vittata formed a close relationship with Ceratopteris richardii with 100% bootstrap support values (). The complete chloroplast (cp) genome of P. vittata will provide valuable molecular data for further phylogenetic studies, as well as fundamental information to survey mechanism of arsenic hyperaccumulation.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cesaro P, Cattaneo C, Bona E, Berta G, Cavaletto M. 2015. The arsenic hyperaccumulating Pteris vittata expresses two arsenate reductases. Sci Rep. 5:14525.
- Christenhusz MJM, Chase MW. 2014. Trends and concepts in fern classification. Ann Bot. 113:571–594.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. https://mafft.cbrc.jp/alignment/software/index.html
- Ruhfel B, Lindsay S, Davis CC. 2018. Phylogenetic placement of Rheopteris and the polyphyly of Monogramma (Pteridaceae s.l.): evidence from rbcL sequence data. Syst Bot. 33:37–43.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. https://sco.h-its.org/exelixis/web/software/raxml/
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Xie ZW. 1996. Compilation of national Chinese herbal medicine. Beijing: People Health Press; p. 646–647. [in Chinese].
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. https://www.ebi.ac.uk/~zerbino/velvet/
- Zhang GM, Liao WB, Ding MY, Lin YX, Wu ZH, Zhang XC, Dong SY, Prado J, Gilbert MG, Yatskievych G, et al. 2013. Pteridaceae. In: Wu ZY, Raven PH, Hong DY, eds. Flora of China. Vol. 2–3 (Pteridophytes). Beijing: Science Press; p. 169–256.