Abstract
The tachinid fly Drino sp. belongs to the subfamily Exoristinae of Tachinidae. We sequenced and annotated the mitogenome of Drino sp. which makes this species first representative of the tribe Eryciini (Tachinidae: Exoristinae) with nearly complete mitochondrial data. This mitogenome is 15437 bp in total, which consists of 22 transfer RNAs, 13 protein-coding genes, 2 ribosomal RNAs and non-coding control region. All genes have the conservational arrangement with other published species of Tachinidae. The nucleotide composition biases toward A and T, the overall A + T% was up to 80.4% of the entire mitogenome. Bayesian inference analysis strongly supported the monophyly of Tachinidae and Exoristinae. Our results also suggested that Exoristinae is the sister group to Phasiinae, and Dexiinae is the sister group to the clade of Phasiinae + Exoristinae.
Among all the species of flies, Tachinidae is one of the most diverse families with over 8,500 known species distributed worldwide. Tachinid flies play an important role in controlling the pests and balancing the ecosystem acting as enemy of pests (Stireman et al. Citation2006; O’Hara et al. Citation2009; O’Hara Citation2016; O’Hara and Cerretti Citation2016). Due to its various morphological features, entomologists face great challenge in identifying tachinid flies, the phylogenetic relationships in Tachinidae also draw the constant attention of the scholars (Meier et al. Citation2006; O’Hara Citation2013; Zhao et al. Citation2013).
Hence, we sequenced the mitochondrial genome of Drino sp., which could be the representative of subfamily Exoristinae for further research. Specimens of Drino sp. were collected in Mt. Wuling of Hebei Province by Kai Wang, and identified by Peng Hou and Prof. Chuntian Zhang. Specimens are deposited in the Entomological Museum of China Agricultural University (CAU).
The genomic DNA was extracted from the adult’s muscle tissues of the thorax using DNeasy DNA Extraction kit (TIANGEN, Beijing, China), and was sequenced under the next generation sequence technology. The mitochondrial genome of Drino sp. contains 22 transfer RNA genes, 13 protein-coding genes (PCGs), two ribosomal RNA genes and one non-coding control region, which were similar with related species reported before (Kang et al. Citation2016; Li et al. Citation2016, Citation2017; Wang et al. Citation2016a, Citation2016b, Citation2016c; Zhao et al. Citation2013).
The nucleotide composition of Drino sp. mitochondrial genome was 41.2% of A, 39.2% of T, 11.6% of C, and 8.0% of G. The A + T content was 80.4%, which was also at an average level among tachinid flies. The codon ATG was the most popular start codon shared with ATP6, CO2, CO3, CYTB, ND4, ND4L, and start codon ATT was shared with ND2, ND3, ND5, ND6. Particularly, the ATP8 begins with codon ATC, the CO1 begins with codon TCG, and the ND1 begins with codon TTG. The conservative stop codon TAA was shared with ATP6, ATP8, CO1, CO3, ND2, ND4L, ND6, another common stop codon TAG was shared with CYTB, ND1, ND3. As for the other three genes, CO2 and ND5 were terminated with an incomplete stop codon T, while the gene ND4 was ended with stop codon TA.
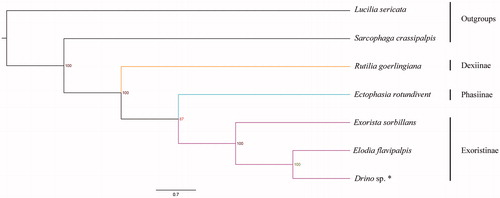
Phylogenetic analysis was performed on the basis of the dataset containing sequences of 13 PCGs from five Tachinidae and two Oestroidea species. The topology provided by Bayesian (BI) analysis based on the PCGs matrices was shown in . According to the phylogenetic outcome, the outgroups Lucilia sericata (Calliphoridae) and Sarcophaga crassipalpis (Sarcophagidae) form a clade diverged from Tachinidae clades. Our results strongly supported the monophyly of Tachinidae as well as subfamily Exoristinae. It also indicated that subfamily Exoristinae is the sister group to subfamily Phasiinae, and subfamily Dexiinae is the sister group to the clade of Phasiinae + Exoristinae.
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Kang Z, Li X, Yang D. 2016. The complete mitochondrial genome of Dixella sp. (Diptera: Nematocera, Dixidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1528–1529.
- Li X, Wang Y, Su S, Yang D. 2016. The complete mitochondrial genomes of Musca domestica and Scathophaga stercoraria (Diptera: Muscoidea: Muscidae and Scathophagidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1435–1436.
- Li X, Ding S, Hou P, Liu X, Zhang C, Yang D. 2017. Mitochondrial genome analysis of Ectophasia rotundiventris (Diptera, Tachinidae). Mitochondrial DNA B. 22:457–458.
- Meier R, Shiyang K, Vaidya G, Ng PKL. 2006. DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst Biol. 55:715–728.
- O’Hara JE, Cerretti P. 2016. Annotated catalogue of the Tachinidae (Insecta, Diptera) of the Afrotropical Region, with the description ofseven new genera. Zookeys. 575:1–344.
- O’Hara JE, Shima H, Zhang C. 2009. Annotated catalogue of the Tachinidae (Insecta: Diptera) of China. Zootaxa. 2190:1–236.
- O’Hara JE. 2013. History of tachinid classification (Diptera, Tachinidae). Zookeys. 31:1–34.
- O’Hara JE. 2016. World genera of the Tachinidae (Diptera) and their regional occurrence. Version 9.0 PDF document. p. 93.
- O’Hara JE, Cerretti P. 2016. Annotated catalogue of the Tachinidae (Insecta, Diptera) of the Afrotropical Region, with the description of seven new genera. Zookeys. 575:1–344.
- Stireman JO, O'Hara JE, Wood DM. 2006. Tachinidae: evolution, behavior, and ecology. Annu Rev Entomol. 51:525–555.
- Wang K, Ding S, Yang D. 2016a. The complete mitochondrial genome of a stonefly species, Kamimuria chungnanshanna Wu, 1948 (Plecoptera: Perlidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:3810–3811.
- Wang K, Wang Y, Yang D. 2016b. The complete mitochondrial genome of a stonefly species, Togoperia sp. (Plecoptera: Perlidae). Mitochondrial DNA A Mapp Seq Anal. 27:1704–3810.
- Wang Y, Liu Y, Yang D. 2016c. The complete mitochondrial genome of a fishfly, Dysmicohermes ingens (Chandler) (Megaloptera: Corydalidae: Chauliodinae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1092–1093.
- Zhao Z, Su T, Chesters D, Wang S, Ho SYW, Zhu C, Chen X, Zhang C. 2013. The mitochondrial genome of Elodia flavipapis Aldrich (Diptera: Tachinidae) and the evolutionary timescale of Tachinid flies. PLoS One. 8:e61814.