Abstract
In the present report, we describe the first sequencing and assembly of the complete mitochondrial genome of Cheilio inermis. The mitochondrial genome of C. inermis, with 16,494 bp in length, has the typical vertebrate mitochondrial gene arrangement. It contains 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a control region. All the tRNA genes typically formed a cloverleaf secondary structure. Phylogenetic analysis using mitochondrial genomes of 11 species showed that C. inermis formed monophyletic group with other Labridae species.
The cigar wrasse, Cheilio inermis (Labriformes: Labridae), is widely distributed throughout the tropical/subtropical Indo-Pacific region, from the Red Sea and East Africa to the Hawaiian and Easter Islands (Randall et al. Citation1990; Cheung et al. Citation2010). This fish inhabits seagrass beds and algal-covered flats, occasionally in lagoon and seaward reefs (Myers Citation1991; Kuiter and Tonozuka Citation2001; Gell and Whittington Citation2002) and is assessed as Least Concern in IUCN Red List (Cheung et al. Citation2010). To our knowledge, this study is the first to determine the sequence of complete mitochondrial genome of C. inermis, and to analyse the phylogenetic relationship of this species with members of Labridae.
The C. inermis specimen used in this study was collected from Chuuk ST 1, Micronesia (7.27N, 151.54E). Total genomic DNA was isolated from tissue of the specimen, which has been deposited in the National Marine Biodiversity Institute of Korea (Voucher No. MABIK 0000619). The mitogenome was sequenced and assembled using Illumina Hiseq 4000 sequencing platform (Illumina, San Diego, CA) and SOAPdenovo assembler at Macrogen Inc. (Seoul, Korea), respectively. The complete mitochondrial genome was annotated using MacClade ver. 4.08 (Maddison and Maddison Citation2005) and DNASIS ver 3.2 (Hitachi Software Engineering, Tokyo, Japan).
The complete mitochondrial genome of C. inermis, with 16,494 bp in length (GenBank accession no. AP018552), and includes 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a control region (D-Loop). The ND6 gene and eight tRNA genes are encoded on the light strand. The overall base composition of the heavy strand is 26.49% A, 30.26% C, 17.42% G, and 25.83% T. Similar to the mitogenomes of other vertebrates, the purine-pyrimidine base pairing of AT content is higher than the GC content (Saccone et al. Citation1999). All tRNA genes can fold into a typical cloverleaf structure, with lengths ranging from 65 to 75 bp. The 12S rRNA (948 bp) and 16S rRNA genes (1,693 bp) are located between tRNAPhe and tRNAVal and between tRNAVal and tRNALeu(UUR), respectively. Of the 13 protein-coding genes, 12 begin with an ATG start codon; the exception being the COI gene, which start with GTG. For the stop codon of the protein-coding genes, five genes ended with a single base T, ATP6 and COIII with TA, ND6 with TAG and NDI, COI, ATP8, ND4L and ND5 with TAA. The control region is 827 bp long and located between tRNAPro and tRNAPhe.
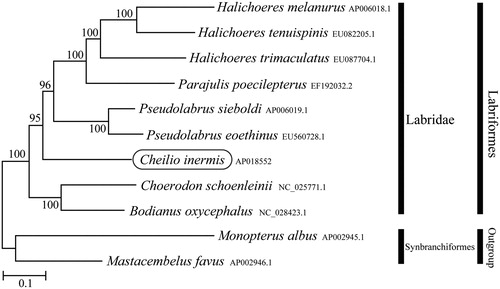
Phylogenetic trees were constructed by the maximum-likelihood method using MEGA 7.0 software (Kumar et al. Citation2016) for the newly sequenced genome and a further 10 complete mitochondrial genome sequences downloaded from the National Center for Biotechnology Information. We confirmed that C. inermis formed a monophyletic group with other Labridae species with high statistical support (). The newly determined mitochondrial genome in this study will provide important DNA molecular data for further phylogenetic analysis and conservation genetics of C. inermis.
Acknowledgements
The authors would like to thank the late Seong-Yong Kim at the National Marine Biodiversity Institute of Korea for collecting the specimen.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cheung WWL, Sadovy Y, Liu M. 2010. Cheilio inermis. The IUCN Red List of Threatened Species 2010: e.T187383A8520063; [accessed 2018 Jun 19]. http://dx.doi.org/10.2305/IUCN.UK.2010-4.RLTS.T187383A8520063.en.
- Gell FR, Whittington MW. 2002. Diversity of fishes in seagrass beds in the Quirimba Archipelago, northern Mozambique. Mar Freshwater Res. 53:115–121.
- Kuiter RH, Tonozuka T. 2001. Pictorial guide to Indonesian reef fishes. Part 2. Fusiliers-Dragonets, Caesionidae-Callionymidae. Zoonetics. Australia.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Maddison DR, Maddison WP. 2005. MacClade 4: analysis of phylogeny and character evolution. ver. 4.08. Sunderland (MA): Sinauer Associates.
- Myers RF. 1991. Micronesian reef fishes. Barrigada (Guam): Coral Graphics.
- Randall JE, Allen GR, Steene RC. 1990. Fishes of the Great Barrier Reef and Coral Sea. Honolulu (HI): University of Hawaii Press.
- Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A. 1999. Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene. 238:195–209.