Abstract
Artemisia selengensis Turcz (Louhao in Chinese) is a widely used health food and a well-known traditional Chinese medicine. However, only a small part of the chloroplast genome data of Artemisia has been reported and there was no report for A. selengensis. In this study, we presented the complete chloroplast genome of A. selengensis and analysed its phylogenetic relationship with other 28 related species belonging to the Asteraceae family. The result showed that the whole genome was 151 215 bp in length with a typical conserved quadripartite structure. The total GC content of whole genome, LSC, SSC, and IRa/IRb regions was 37.46, 35.55, 30.81, and 43.09%, respectively. A total of 133 genes were identified, including 88 protein-coding, 37 transfer RNA, and eight ribosome RNA genes. Among these genes, nineteen genes contained a single intron and two genes contained two introns. The phylogenetic relationship showed that A. selengensis was similar to A. gmelinii. The complete chloroplast genome presented here will enrich the genetic resources of medicinal plant and promote our understanding of the phylogeny of Artemisia within the Asteraceae family.
Artemisia selengensis Turcz (Louhao in Chinese) which belongs to the Asteraceae family is a widely used health food for its taste and nutritional properties. It has also been used as a well-known traditional Chinese medicine because some extracted substances from it have potent antitumor, antioxidant, and free radical scavenging activities (Koo et al. Citation1994; Shi et al. Citation2010). Actually, except for A. selengensis, many species of Artemisia L. are used as herbal medicines in many countries (Tariq et al. Citation2011). In result, the genus Artemisia has been the subject of genetic diversity or phylogeny studies. Previous studies have been conducted on phylogenetic relationships within Artemisia by using chloroplast DNA data, like psbA-trnH makers (Haghighi et al. Citation2014), rps11 genes (Tariq et al. Citation2011), and ribosomal and chloroplast sequences (Pellicer and Garnatje Citation2011). However, only a fraction of the chloroplast genome data of Artemisia has been reported and there was no report for A. selengensis. Therefore, we presented the complete chloroplast genome of A. selengensis to enrich the genetic resources of medicinal plant and to have a comprehensive knowledge on the phylogeny of Artemisia.
The mature leaves of A. selengensis were collected from a natural population in Dongting Lake region (28°35′6.29″N and 112°20′6.29″E) and stored at −80 °C in Hunan Research Center of Engineering Technology for Utilization of Environmental and Resources Plant with accession number 20171105AS. The total chloroplast DNA was extracted by using Plant Chloroplast Purification Kit (BTN120308) and Column Plant DNA Extraction Kit and sequenced by using Illumima Hiseq 4000 Platform with Zhang’s method (Zhang et al. Citation2017). The raw data was filtered using Trimmomatic version 0.32 (Aachen, Germany, Bolger et al. Citation2014) to obtain clean data for subsequent analysis. The annotation of the whole genome and drawing of a circular map were generated by using DOGMA (Wyman et al. Citation2004) and OGDRAW (Lohse et al. Citation2007), respectively. The complete chloroplast genome was submitted to the NCBI database under the accession number MH042532.
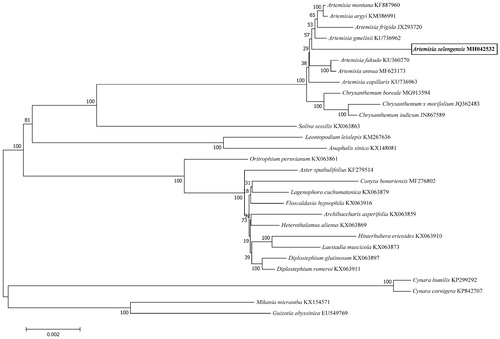
The complete chloroplast genome of A. selengensis which contained a typical conserved quadripartite structure, with a LSC region of 82 920 bp, a SSC region of 18 367 bp, and a pair of IRs regions of 24 964 bp, was 151 215 bp in length. The total GC content of whole genome, LSC, SSC, and IRa/IRb regions was 37.46, 35.55, 30.81, and 43.09%, respectively. A total of 133 genes were identified, including 88 protein-coding, 37 transfer RNA, and eight ribosome RNA genes. Among these genes, nineteen genes (rps16, rpoC1, atpF, petB, petD, rpl16, rpl2 × 2, ndhB × 2, ndhA, ycf3, clpP, trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU × 2, and trnA-UGC × 2) contained a single intron and two genes (ycf3 and clpP) contained two introns. We further analysed phylogenetic relationship of A. selengensis with other 28 related species belonging to the Asteraceae family based on 72 shared protein-coding sequence by using neighbour-joining (NJ) method in MEGA version 7.0 (Arizona, USA, ) (Kumar et al. Citation2016). Our result confirmed that A. selengensis was similar to A. gmelinii. In all, the complete chloroplast genome presented here will enrich the genetic resources of medicinal plant and promote our understanding of the phylogeny of Artemisia within the Asteraceae family.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Haghighi AR, Belduz AO, Vahed MM, Coskuncelebi K, Terzioglu S. 2014. Phylogenetic relationships among Artemisia species based on nuclear ITS and chloroplast psbA-trnH DNA markers. Biologia. 69:834–839.
- Koo KA, Kwak JH, Lee KR, Zee OP, Woo ER, Park HK, Youn HJ. 1994. Antitumor and immunomodulating activities of the polysaccharide fractions from Artemisia selengensis and Artemisia iwayomogi. Arch Pharm Res. 17:371–374.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52:267–274.
- Pellicer J, Garnatje T. 2011. Phylogenetic relationships of Artemisia subg. Dracunculus (Asteraceae) based on ribosomal and chloroplast DNA sequences. Taxon. 60:691–704.
- Shi F, Jia X, Zhao C, Chen Y. 2010. Antioxidant activities of various extracts from Artemisisa selengensis Turcz (LuHao). Molecules. 15:4934–4946.
- Tariq M, Nadia H, Nazia N, Ishrat N. 2011. Phylogenetic analysis of different Artemisia species based on chloroplast gene rps11. Arch Biol Sci. 63:661–665.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhang W, Zhao YL, Yang GY, Tang YC, Xu ZG. 2017. Characterization of the complete chloroplast genome sequence of Camellia oleifera in Hainan, China. Mitochondrial DNA. 2:843–844.