Abstract
Mesopteris tonkinensis is monotypic species in the genus Mesopteris (Thelypteridaceae). We characterized its complete chloroplast genome sequences by Illumina sequencing and de novo assembly. The genome size is 161,380 bp in length with a GC content of 43.6%, containing a large single-copy (LSC) region of 82,678 bp, a small single-copy (SSC) region of 21,786 bp and a pair of inverted repeat (IR) of 28,458 bp. In total, 131 genes are identified, including 88 protein-coding genes, 34 tRNA genes with absent trnV-UAC, eight rRNA genes and one pseudogene. ML tree revealed that M. tonkinensis and Christella appendiculata were closely related.
Mesopteris tonkinensis is monotypic species in the genus Mesopteris (Thelypteridaceae) (Lin et al. Citation2013). It is endemic to wet rocks in limestone areas in South China and North Vietnam (Ching Citation1978). The feature of this fern reflects in callose protuberance at the sinuses and 2½ pairs of veinlets connivent under the sinuses (He and Zhang Citation2012). Established since 1934, M. tonkinensis was successively ascribed to Dryopteris (Dryopteridaceae), Thelypteris, Lastrea and Amphineuron (Thelypteridaceae) due to its unique characteristics (Wang et al. Citation2015). The first molecular phylogenetic analysis strongly supported M. tonkinensis as monophyletic and as a part of the Cyclosorus clade (He and Zhang Citation2012). Because of its inaccessibility, the research of M. tonkinensis is only limited in morphology, cytology and phylogeny (Huang and Zhou Citation1994; He and Zhang Citation2012; Wang et al. Citation2015). Hence, it is necessary to acquire whole chloroplast (cp) genome sequence of M. tonkinensis, which provide more useful information to determine its classification position.
A plant material of M. tonkinensis was provided by South China Botanical Garden, Chinese Academy of Sciences (23°11′3.56″N, 113°21′43.28″E) and saved in Herbarium of Sun Yat-sen University (SYS; voucher: SS Liu 201617). Genomic DNA was extracted from fresh leaves by Tiangen Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China) and broken into 300 bp with Covaris M220 (Covaris Inc., MS, USA). A paired-end (PE) library was constructed using NEBNext Ultra DNA Library Prep Kit for Illumina (New England BioLabs Inc., Ipswich, MA). After sequencing was performed in Illumina Hiseq 2500 platform (Illumina Inc., San Diego, USA), we removed the adapters and low-quality reads from 2.38G raw data through Trimmomatic v0.32 (Bolger et al. Citation2014) and qualified 2.05G clean data by FastQC v0.10.0 (Andrews Citation2010). The complete chloroplast genome was de novo assembled by Velvet v1.2.07 (Zerbino and Birney Citation2008), and further annotated using DOGMA (Wyman et al. Citation2004) and tRNAscan-SE (Schattner et al. Citation2005), and finally manual confirmation based on BLAST searches. In order to further analyze phylogenetic relationships, ten ferns including Ophioglossum californicum as outgroup were selected and aligned with MAFFT v.7.221 (Katoh and Standley Citation2013). Phylogenetic tree was generated by a maximum likelihood analysis of RAxML v8.0 (Stamatakis Citation2014) with 1000 bootstrap replicates.
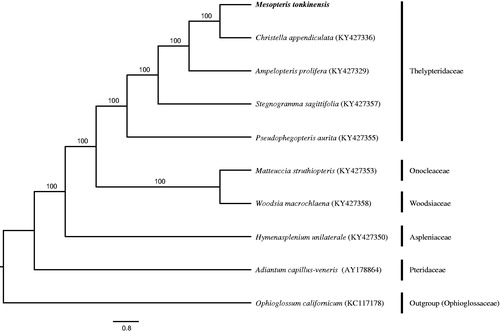
The complete chloroplast genome of M. tonkinensis is a typical quadripartite structure with 161,380 bp, which was separated into a large single-copy (LSC) region of 82,678 bp, a small single-copy (SSC) region of 21,786 bp and a pair of inverted repeat (IR) regions of 28,458 bp (GenBank accession number: MH500229). Whole genome presents the GC content of 43.6%, whereas corresponding GC value in LSC, SSC, and IR is 42.2%, 39.87%, and 46.9%, respectively. The genome contains 88 protein-coding genes, 34 tRNA genes with absent trnV-UAC, eight rRNA genes and one pseudogene (ndhB). Among these genes, 116 genes are single-copy genes, 15 genes (ndhB, rps16, atpF, rpoC1, petB, petD, ndhA, rpl16, rpl2, trnG-UCC, trnV-UAC, trnA-UGC, trnI-GAU, trnL-UAA, and trnT-UGU) harbor a single intron, and three genes (ycf3, clpP, and rps12) have two introns. ML tree revealed that M. tonkinensis and Christella appendiculata were closely related with high bootstrap support values ().
Disclosure statement
The authors declare no conflict of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Cambridge (UK): the Babraham Institute; [accessed 2017 Jul 29]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Ching RC. 1978. The Chinese fern families and genera: systematic arrangement and historical origin. Acta Phytotax Sin. 16:1–19.
- Huang YY, Zhou HG. 1994. Anatomical study on Mesopteris tonkinensis. J Guangxi Agri Uni. 13:204–210.
- He LJ, Zhang XC. 2012. Exploring generic delimitation within the fern family Thelypteridaceae. Mol Phylogenet Evol. 65:757–764.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Lin YX, Li ZY, Iwatsuki K, Smith AR. 2013. Thelypteridaceae. In: Wu ZY, Raven PH, Hong DY, eds., Flora of China. Vol. 2-3 (Pteridophytes). Beijing: Science Press; p. 319–396.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–689.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Wang RX, Shao W, Liu L, Liu J, Deng XC, Lu SG. 2015. A systematic study of the fern genus Mesopteris Ching (Thelypteridaceae). Am Fern J. 105:11–19.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.