Abstract
Lonicera japonica is a traditional medicinal plant well known for its anti-inflammatory effect. The complete chloroplast genome sequence of L. japonica collected from Korea was obtained by de novo assembly using whole genome sequence data. The chloroplast genome is 155,060 bp in length, containing 88,853 bp in a large single copy (LSC), 18,653 bp in a small single copy (SSC) and 23,777 bp in a pair of inverted repeats (IRs). A total of 112 genes including 78 protein-coding genes and 34 structural RNA genes were identified. The sequence comparison of two L. japonica collected from Korea and China revealed 48 single nucleotide polymorphisms (SNPs) and 45 insertions/deletions (InDels). In addition, phylogenetic analysis represented intraspecific diversity within L. japonica species collected in Korea and China.
The Lonicera genus belongs to the Caprifoliaceae family which is close to the Rubiaceae family and consists of approximately 200 species that are mainly distributed in East Asia. Among them, about 100 inhabit in China, about 25 in Japan and about 30 in Korea. Many Lonicera species have been used as herbal medicines. L. japonica, called golden-and-silver honeysuckle or Japanese honeysuckle, has been widely used in traditional herbal medicine (Peng et al. Citation2000), and its flower bud also has been prescribed to treat some infectious diseases due to its anti-inflammatory and antiviral effects (Chang and Hsu Citation1992). These effects come from many active compounds identified in the stems and leaves of L. japonica (Shang et al. Citation2011). Previously, a chloroplast genome sequence of L. japonica collected from China (China collection) has been reported (He et al. Citation2017). In this study, we characterized the complete chloroplast genome sequence of L. japonica collected from Korea (Korea collection) and compared it with the Chinese L. japonica chloroplast genome.
The plant sample was collected from Medicinal Plant Garden, College of Pharmacy, Seoul National University, Koyang, Korea. Total genomic DNA was extracted from fresh leaves using a modified cetyltrimethylammonium bromide protocol (Allen et al. Citation2006) and sequenced by the Illumina MiSeq platform (Illumina, San Diego, CA). Whole genome sequence data of 4.1 Gb in paired-end reads were obtained and assembled de novo using CLC genome assembler (v. beta 4.6, CLC Inc., Aarhus, Denmark) as previously described (Kim et al. Citation2015a). The gene annotation of the complete chloroplast genome was performed using GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html), followed by manual confirmation using Artemis programme (Rutherford et al. Citation2000) and BLAST searches.
Complete chloroplast genome of L. japonica (Korea collection) was 155,060 bp in length, and separated into four distinct regions: a large single copy (LSC) (88,853 bp), a small single copy (SSC) (18,653 bp), and a pair of inverted repeats (IRs) (each 23,777 bp). In Korea J. japonica collection chloroplast genome, a total of 112 genes including 78 protein-coding genes, 30 tRNA genes, and 4 rRNA genes were identified.
Chloroplast genome comparison between Korea and China L. japonica collections revealed that the Korea collection was 18 bp shorter than China collection, 5 bp and 19 bp shorter in LSC and SSC regions, respectively, but 3 bp longer in each IR region. We identified 48 SNPs and 45 InDels between these two L. japonica collections that show richer intra-species diversity such as Panax ginseng and Brassica species (Kim et al. Citation2015b, Citation2018; Joh et al. Citation2017; Nguyen et al. Citation2017, Citation2018).
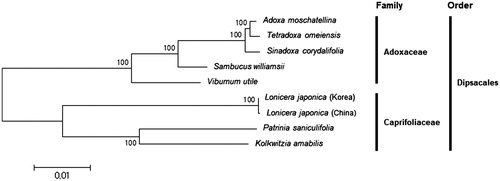
A phylogenetic tree based on complete chloroplast genome sequences was constructed with seven other species including two in Caprifoliaceae and five in Adoxaceae (). As expected, Korea and China collections of L. japonica were clustered together and grouped with other Caprifoliaceae species. Based on the branch lengths, L. japonica (China collection) has a long branch and it is thought more diversified from their common ancestor than L. japonica (Korea collection).
Figure 1. Phylogenetic tree of Lonicera and other related species in Dipsacales. The tree was constructed using complete chloroplast genome sequences of the nine species and analysed by the neighbour-joining method with 1000 bootstrap values in MEGA 6.0 (Tamura et al. Citation2013). The numbers in the nodes indicate bootstrap support values. The chloroplast genome sequences used for this tree are: Adoxa moschatellina, NC_034792.1; Kolkwitzia amabili, NC_029874.1; Lonicera japonica (Korea), MH028738; Lonicera japonica (China), NC_026839.1; Patrinia saniculifolia, NC_036835.1; Sambucus williamsii, NC_033878.1; Sinadoxa corydalifolia, NC_032040.1; Tetradoxa omeiensis, NC_034793.1; Viburnum utile, NC_032296.1.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Allen G, Flores-Vergara M, Krasynanski S, Kumar S, Thompson W. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1:2320.
- Chang WC, Hsu FL. 1992. Inhibition of platelet activation and endothelial cell injury by polyphenolic compounds isolated from Lonicera japonica Thunb. Prostaglandin Leukotriene Essent Fatty Acids. 45:307–312.
- He L, Qian J, Li X, Sun Z, Xu X, Chen S. 2017. Complete chloroplast genome of medicinal plant lonicera japonica: genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules. 22:249.
- Joh HJ, Kim NH, Jayakodi M, Jang W, Park JY, Kim YC, In JG, Yang TJ. 2017. Authentication of golden-berry P. ginseng cultivar ‘gumpoong’ from a landrace ‘hwangsook’ based on pooling method using chloroplast-derived markers. Plant Breed Biotechnol. 5:16–24.
- Kim CK, Seol YJ, Perumal S, Lee J, Waminal NE, Jayakodi M, Lee SC, Jin S, Choi BS, Yu Y. 2018. Re-exploration of U’s triangle brassica species based on chloroplast genomes and 45S nrDNA sequences. Sci Rep. 8:7353.
- Kim K, Lee S, Lee J, Yu Y, Yang Y, Choi B, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015a. Complete chloroplast ad ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ. 2015b. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One. 10:e0117159.
- Nguyen VB, Park HS, Lee SC, Lee J, Park JY, Yang TJ. 2017. Authentication markers for five major Panax species developed via comparative analysis of complete chloroplast genome sequences. J Agric Food Chem. 65: 6298–6306.
- Nguyen, VB, Giang VNL, Waminal NE, Park HS, Kim NH, Jang W, Lee J, Yang TJ. 2018. Comprehensive comparative analysis of chloroplast genomes from seven Panax species and development of an authentication system based on species-unique SNP markers. J Ginseng Res.
- Peng LY, Mei SX, Jiang B, Zhou H, Sun HD. 2000. Constituents from Lonicera japonica. Fitoterapia. 71:713–715.
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics. 16:944–945.
- Shang X, Pan H, Li M, Miao X, Ding H. 2011. Lonicera japonica Thunb: ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. J Ethnopharmacol. 138:1–21.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
