Abstract
The complete mitochondrial genome of a freshwater planorbid snail, Planorbella duryi (Mollusca, Gastropoda) was recovered from de novo assembly of genomic sequences generated with the Illumina NextSeq500 platform. The P. duryi mitogenome (14,217 base pairs) is AT rich (72.69%) and comprises 13 protein-coding genes, two ribosomal subunit genes, and 22 transfer RNAs. The gene order is identical to that of Biomphalaria glabrata and other snail species in the family Planorbidae. This is the first full characterization of a mitochondrial genome of the genus Planorbella.
Keywords:
Report
Mitogenome data have benefited phylogenetic reconstructions of the class Gastropoda (phylum Mollusca), a group with a complex taxonomy (White et al. Citation2011; Williams et al. Citation2014; Oskars et al. Citation2015). This report presents the mitochondrial genome for the snail Planorbella duryi adding to the limited availability of mitogenome data available for the family Planorbidae that includes several medically important snails that transmit schistosome parasites of humans. (DeJong et al. Citation2004; Jannotti-Passos et al. Citation2010; Zhang et al. Citation2018).
An individual snail, morphologically identified as a planorbid, was collected from Shady lakes trout fishing resort in Albuquerque, NM (35.2144° N, 106.5956° W). After removal of the shell, genomic DNA was extracted from whole-body tissues using a CTAB-based method (Winnepenninckx et al. Citation1993). High-throughput DNA sequencing was carried out using the KAPA Hyper Prep Kit, Illumina® platforms (KAPPA Biosystems) and the Illumina NextSeq500 at the Molecular Biology Facility of UNM. The whole body tissues of P. duryi were destructively sampled for DNA, however, all genomic Illumina data were deposited in the SRA database (GenBank accession SRP151592). The mitochondrial genome and two nuclear genes (18S, 28S) were assembled using MITObim v 1.8 (Hahn et al. Citation2013). MITOS (Bernt et al. Citation2013) annotation of the assembled mitogenome was manually checked to minimize overlaps of protein-coding genes and identify candidate stop codons that are completed by polyadenylation (DeJong et al. Citation2004). Protein-coding (amino acid) sequences of complete mitogenomes of P. duryi (KY514384) and select gastropod taxa were aligned using ClustalX (Larkin et al. Citation2007) and trimmed to minimize gaps at sequence termini. Phylogenetic reconstruction was performed using MEGA v.7 (Kumar et al. Citation2016) and Neighbor-Joining statistical method with 1,000 bootstrap replicates (JTT model with gamma distributed rate variation among sites).
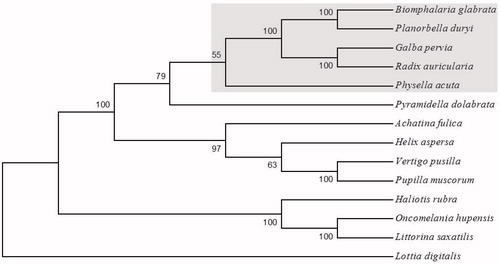
The species designation Planorbella duryi was consistent with snail morphology, knowledge of local snail fauna, and BLAST analyses of 18S and 28S sequences (GenBank accessions KY514382 and KY514383). The complete mitochondrial genome of P. duryi is 14,217 bp in length (GenBank accession: KY514384) and contains 13 protein-coding genes, two ribosomal subunit genes (12S, 16S), and 22 tRNAs. Similar to other gastropod species, the P. duryi mitogenome is AT rich with a base composition of 31.01% A, 13.03% C, 14.29% G, and 41.68% T. Illustrative of typical panpulmonate snails (excluding Physella acuta; Nolan et al. Citation2014), the gene order of the P. duryi mitogenome is identical to that of the other species in GenBank, including Biomphalaria snails, close relatives within the family Planorbidae (DeJong et al. Citation2004). Additionally, phylogenetic analyses confirmed P. duryi as a sister species to B. glabrata, within the family Planorbidae of the Hygrophila (). The characterization of the mitogenome of P. duryi can help elucidate challenging phylogenetic reconstruction of Gastropoda and provide a genomic resource for continued research on snails and their role in transmission of the infectious disease schistosomiasis.
Figure 1. Neighbor-joining tree of complete mitogenomes (protein-coding sequences) of Planorbella duryi and 13 gastropod species. Planorbella duryi is placed among species of the taxonomic clade Hygrophila (shaded). The basal gastropod Lottia digitalis is used as an outgroup. Accessions for full mitogenome sequences: Achatina fulica (NC_024601), Biomphalaria glabrata (NC_005439), Galba pervia (NC_018536), Haliotis rubra (NC_005940), Helix aspersa (NC_021747), Littorina saxatilis (NC_030595), Lottia digitalis (NC_007782), Oncomelania hupensis (NC_013073), Physella acuta (NC_023253), Planorbella duryi (KY514384), Pupilla muscorum (NC_026044), Pyramidella dolabrata (NC_012435), Radix auricularia (NC_026538), Vertigo pusilla (NC_026045).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- DeJong RJ, Emery AM, Adema CM. 2004. The mitochondrial genome of Biomphalaria glabrata (Gastropoda: Basommatophora), intermediate host of Schistosoma mansoni. J Parasitol. 90:991–997.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nuc Acids Res. 41:e129.
- Jannotti-Passos LK, Ruiz JC, Caldeira RL, Murta SM, Coelho PM, Carvalho OS. 2010. Phylogenetic analysis of Biomphalaria tenagophila (Orbigny, 1835) (Mollusca: Gastropoda). Mem Inst Oswaldo Cruz. 105:504–511.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948.
- Nolan JR, Bergthorsson U, Adema CM. 2014. Physella acuta: atypical mitochondrial gene order among panpulmonates (Gastropoda). J Molluscan Stud. 80:388–399.
- Oskars TR, Bouchet P, Malaquias MA. 2015. A new phylogeny of the Cephalaspidea (Gastropoda: Heterobranchia) based on expanded taxon sampling and gene markers. Mol Phylogenet Evol. 89:130–150.
- White TR, Conrad MM, Tseng R, Balayan S, Golding R, de Frias Martins AM, Dayrat BA. 2011. Ten new complete mitochondrial genomes of pulmonates (Mollusca: Gastropoda) and their impact on phylogenetic relationships. BMC Evol Biol. 11:295.
- Williams ST, Foster PG, LIttlewood DT. 2014. The complete mitochondrial genome of a turbinid vetigastropod from MiSeq Illumina sequencing of genomic DNA and steps towards a resolved gastropod phylogeny. Gene. 533:38–47.
- Winnepenninckx B, Backeljau T, De Wachter R. 1993. Extraction of high molecular weight DNA from molluscs. Trends Genet. 9:407.
- Zhang S-M, Bu L, Laidemitt MR, Lu L, Mutuku MW, Mkoji GM, Loker ES. 2018. Complete mitochondrial and rDNA complex sequences of important vector species of Biomphalaria, obligatory hosts of the human-infecting blood fluke, Schistosoma mansoni. Sci Rep. 8:7341.
