Abstract
Styrax zhejiangensis is an endemic species to China and is only distributed in Jiande, Zhejiang Province. The species is on the verge of extinction. The chloroplast genome of S. zhejiangensis was determined from Illumina pair-end sequencing data. The sequence was 157,387 bp and consisted of one large (LSC, 87,193 bp) and one small (SSC, 18,286 bp) single-copy region region, separated by a pair of inverted repeat (IR, 25,954 bp) regions. The sequence included 116 genes, including 82 protein-coding genes, 19 rRNAs and 15 tRNAs. The overall GC content was 37.0%. A maximum likelihood phylogenetic analysis showed that Styracaceae was more closely related to Symplocaceae than to Ebenaceae.
Styrax zhejiangensis, one of 234 Styracaceae species, is a broad-leaved tree species with beautiful white flowers (Wu et al., Citation2013). S. zhejiangensis is endemic to Zhejiang Province, China, with an extremely narrow distribution area. At present, only one very small population, with less than 70 adult individuals of S. zhejiangensis, has been found at the Jiande Forest Farm. The species is on the verge of extinction. There is not any report of this Endangered species in conservation biology and phylogeny studies, except revisions of some species in the same genus (Gene Citation1974, Peter Citation1997, Citation1999). Chloroplast genomes are widely used in studies of species conservation, genome evolution, and phylogeny (Jansen et al. Citation2007, Moore et al. Citation2007). Based on the complete chloroplast genome sequence of S. zhejiangensis, analysis of the composition and structure of this chloroplast genome can reveal more information about its genetic variation, eliciting mechanism.
We sampled a single individual of S. zhejiangensis for total genomic DNA from the nursery of the Zhejiang Academy of Forestry, China, located at 30.21°N, 120.02°E. High-throughput sequencing was carried out using HiSeq™ 2000 analyzer (Illumina, San Diego, California). All the data were assembled into the genome using SPAdes v3.9.0 (Bankevich et al. Citation2012). The annotation was performed with the DOGMA (Wyman et al. Citation2004). Protein-coding genes were identified using the plastid/bacterial genetic code. Intron/exon boundaries were determined using the MAFFT v7 (Katoh and Standley Citation2013), and tRNA boundaries were verified using tRNAscan-SE with default settings (Schattner et al. Citation2005). The GC content was measured using Geneious version 8.0.2 (Kearse et al. Citation2012).
The complete chloroplast genome of S. zhejiangensis (GenBank accession number MG702338, https://www.ncbi.nlm.nih.gov/nuccore/MG702338.1/) was 157,387 bp in length, and the circle was typical in its general structure of most plastids with a pair of IRS (25,954 bp) which were separated by a large single-copy region (LSC; 87,193 bp), and a small single-copy region (SSC region; 18,286 bp). The genome encoded 116 functional genes, consisting of 82 protein-coding genes, 19 ribosomal RNA genes, and 15 transfer RNA genes. Among these genes, 15 genes harbored a single intron. 18 genes were duplicated in the IR region. The overall GC content was 37.0%, while the corresponding values of the of IR, LSC, and SSC regions were 42.5, 34.8, and 30.3%, respectively.
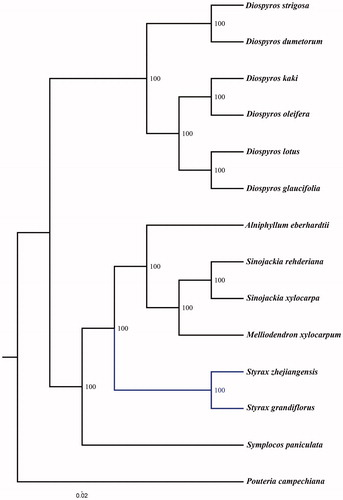
To ascertain the phylogenetic placement of S. zhejiangensis within the order Ebenales, maximum likelihood analysis was performed using the MEGA6 software (Tamura et al. Citation2013) with 1000 bootstrap replicates, according to chloroplast genomes of 14 related species with Pouteria campechiana as an outgroup. The tree revealed that six Styracaceae species formed a monophyletic clade with 100% bootstrap support,and Sinojackia was most closely related to Melliodendron and nearer to Alniphyllum than to Styrax consisting of S. zhejiangensis and S. grandiflora. Styracaceae was more closely related to Symplocaceae than to Ebenaceae (). This newly characterized complete chloroplast genome can provide essential resources for the study of genetic diversity and the conservation of this endangered species.
Figure 1. Phylogenetic tree based on 14 complete chloroplast genome sequences. Accession numbers: Styrax grandiflorus (KX111381.1), Alniphyllum eberhardtii (KX765434.1), Melliodendron xylocarpum (MF179500.1), Sinojackia xylocarpa (KY709672.1), Sinojackia rehderiana (MF179499.1), Diospyros lotus (KM522849.1), Diospyros glaucifolia (KM504956.1), Diospyros kaki (KT223565.1), Diospyros oleifera (KM522850.1), Diospyros dumetorum (MF179487.1), Diospyros strigosa (MF179495.1), Pouteria campechiana (KX426215.1), Symplocos paniculata (MF179486.1).
This work was supported by the Special Fund for Forestry Development and Resources Protection (No. 201711) of Forestry Department of Zhejiang Province, China.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Gene JG. 1974. A revision of Styrax (Styracaceae) in north America, Central America, and the Caribbean. SIDA, Contributions to Botany. 5:191–258.
- Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, Muller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifes genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 104:19369–19374.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Moore MJ, Bell CD, Soltis PS, Soltis DE. 2007. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci USA. 104:19363–19368.
- Peter WF. 1997. A revision of Styrax (Styracaceae) for Western Texas, Mexico, and Mesoamerica. Ann Mo Bot Gard. 84:705–761.
- Peter WF. 1999. Phylogeny of Styrax based on morphological characters, with implications for biogeography and infrageneric classification. Syst Bot. 24:356–378.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0.Mol. Biol Evol. 30:2725–2729.
- Wu ZH, Raven PH, Hong DY.. 2013. Flora of China. Beijing: Science Press, 257 p.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
