Abstract
We analyzed the complete mitochondrial genome of the dusky brown-gray–colored honeybee Apis mellifera, collected from North Island, New Zealand. We determined that the mitochondrial genome was a 16,336 bp and predicted 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes. The start codon ATA was found in two genes, ATG in four genes, ATT in six genes, and ATC in one gene, whereas the termination codon TAA was observed in all PCGs. The non-coding regions of tRNA-Leu and COII were consistent with the C haplotype of A. mellifera carnica. Phylogenetic analysis suggests a close relationship with the European A. mellifera.
The western honeybee, Apis mellifera, is naturally distributed in Africa, the Middle East, and Europe, and is exported worldwide. In New Zealand, the European honeybee subspecies A. m. mellifera was first introduced from the UK in 1839 (Beard Citation2015). Later, two subspecies, A. m. carnica and A. m. ligustica, were introduced to New Zealand from Europe and Australia. Import of honeybees is now prohibited to control the spread of disease (Beard Citation2015). Currently, there are three types of honeybees in New Zealand, each with different body colors. Yellow-colored honeybees (A. m. ligustica) have an Italian origin, as shown by the analysis of complete mitochondrial DNA (Nakagawa et al. Citation2018). Here, we report the complete mitochondrial genome sequence of the New Zealand dusky brown-gray–colored honeybee, A. mellifera. This taxon provides an important sample for rigorous phylogenetic studies of New Zealand honeybees and other A. mellifera subspecies.
Adult workers were collected in March 2017 from Kaitaiya, North Island, New Zealand (−35°11′45, 173°25′39). Genomic DNA was isolated from one worker and sequenced using Illumina’s HiSeq platform (Illumina Inc., San Diego, CA). We generated a DNA library of individuals from New Zealand that consisted of 1,879,764 reads. The resultant reads were assembled and annotated using the MITOS web server (Germany; Bernt et al. Citation2013) and Geneious R9 (Biomatters, New Zealand). A phylogenetic tree was constructed using MEGA6 (Tamura et al. Citation2013) and TREEFINDER (Jobb Citation2015) using the nucleotide sequences of 13 protein-coding genes (PCGs). The DNA specimen was stored in the National Museum of Nature and Science, Japan, accession number: NSMT-I- HYM 75324.
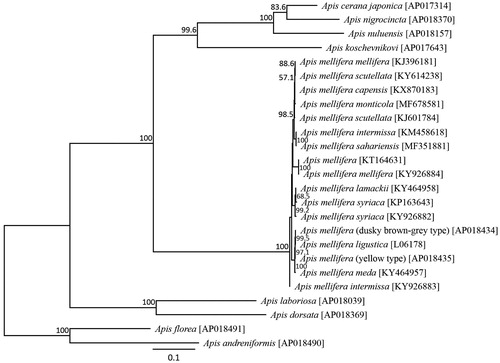
The dusky brown-gray–colored A. mellifera mitochondrial genome was determined to be 16,336 bp long (AP018434). This size is typical for a hymenopteran species. It was found to be similar to the common A. mellifera mitochondrial genome organization, comprising 13 PCGs, 22 putative tRNA genes, two rRNA genes, and an A + T-rich control region. The average AT content of the A. mellifera mitochondrial genome was 84.95%. Similar to the honeybee mitochondrial genomes, the heavy strand encoded nine protein-coding genes and 14 tRNA genes, and the light strand encoded four protein-coding genes, eight tRNAs, and two rRNA genes. ATP6 and ATP8 shared 19 nucleotides in common. Six protein-coding genes of the A. mellifera mitochondrial genome started with ATT, while ATP6, COIII, ND4 and Cytb started with ATG, and COI and ND3 started with ATA. ND2 stared with ATC. This pattern is commonly found in A. mellifera subspecies (Crozier and Crozier Citation1993; Gibson and Hunt Citation2015; Haddad Citation2015; Hu et al. Citation2015; Eimanifar et al. Citation2016a, 2016b, Citation2017a, 2017Citationb, 2017c; Haddad et al. Citation2017; Nakagawa et al. Citation2018). The stop codon of these genes was uniformly TAA, similar to other honeybee subspecies. All the tRNA genes possessed inferred cloverleaf secondary structures, except for Gln, Ser1, and Thr, which lacked the arm structure. Phylogenetic analysis was conducted using 13 mitochondrial PCG sequences from 24 closely related taxa (). The New Zealand dusky brown-gray–colored A. mellifera was found to be most closely related to the European honeybee.
Figure 1. Inferred phylogenetic relationships of the genus Apis (Hymenoptera) using mitochondrial sequences from 13 protein-coding genes under the maximum likelihood criterion. Numbers at the nodes indicate bootstrap support (1,000 replicates). A. florea, A. andreniformis, A. dorsata, A. laboriosa, A. cerana, A. nigrocincta, and A. koschevnikovi (Takahashi et al. Citation2016, Citation2017a, Citation2017b, Citation2017c, Citation2017d, Citation2018; Wakamiya et al. Citation2017) were used as an outgroup. The alphanumeric terms in the parentheses indicate the GenBank accession numbers.
Acknowledgments
We are grateful to Dr. T. Kiyoshi, Mr. Kano and Mr. N. Raymond for valuable comments on the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Beard C. 2015. Honeybees (Apis mellifera) on public conservation lands: a risk analysis. Department of Conservation, Wellington.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo Metazoan Mitochondrial Genome Annotation. Mol Phyl Evol. 69:313–319.
- Crozier RH, Crozier YC. 1993. The mitochondrial genome of the honeybee Apis mellifera: Complete sequence and genome organization. Genetics. 133:97–117.
- Eimanifar A, Kimball RT, Braun EL, Ellis JD. 2016a. The complete mitochondrial genome of the Cape honey bee, Apis mellifera capensis Esch. (Insecta: hymenoptera: apidae). Mitochondrial DNA. 1:817–819.
- Eimanifar A, Kimball RT, Braun EL, Ellis JD. 2016b. The complete mitochondrial genome of the hybrid honey bee, Apis mellifera capensis × Apis mellifera scutellata, from South Africa. Mitochondrial DNA. 1:856–857.
- Eimanifar A, Kimball RT, Braun EL, Fuchs S, Grünewald B, Ellis JD. 2017a. The complete mitochondrial genome of Apis mellifera meda (Insecta: Hymenoptera: Apidae). Mitochondrial DNA. 2:589–269.
- Eimanifar A, Kimball RT, Braun EL, Fuchs S, Grünewald B, Ellis JD. 2017b. The complete mitochondrial genome of an east African honey bee, Apis mellifera monticola Smith (Insecta: Hymenoptera: Apidae). Mitochondrial DNA. 2:589–590.
- Eimanifar A, Kimball RT, Braun EL, Moustafa DM, Haddad N, Fuchs S, Grünewald B, Ellis JD. 2017c. The complete mitochondrial genome of the Egyptian honey bee, Apis mellifera lamarckii (Insecta: Hymenoptera: Apidae). Mitochondrial DNA. 2:589–272.
- Gibson JD, Hunt GJ. 2015. The complete mitochondrial genome of the invasive Africanized Honey Bee, Apis mellifera scutellata (Insecta: Hymenoptera: Apidae). Mitochondrial DNA. 27:561–562.
- Haddad NJ, Adjlane N, Loucif-Ayad W, Dash A, Naganeeswaran S, Rajashekar B, Al-Nakkeb K, Sicheritz-Ponten T. 2017. Mitochondrial genome of the North African Sahara Honeybee, Apis mellifera sahariensis (Hymenoptera: Apidae). Mitochondrial DNA. 2:548–549.
- Haddad NJ. 2015. Mitochondrial genome of the Levant Region honeybee, Apis mellifera syriaca (Hymenoptera: Apidae). Mitochondrial DNA. 30:1–2.
- Hu P, Lu Z-X, Haddad N, Noureddine A, Loucif-Ayad Q, Qang YZ, Zhao R-B, Zhang A-L, Guan X, Zhang H-X, Niu H. 2015. Complete mitochondrial genome of the Algerian honeybee, Apis mellifera intermissa (Hymenoptera: Apidae). Mitochondrial DNA. 26:1–2.
- Jobb G. 2015. TREEFINDER. http://www.treefinder.de/index_treefinder.html
- Nakagawa I, Maeda M, Chikano M, Okuyama H, Murray R, Takahashi J. 2018. The complete mitochondrial genome of the yellow colored honeybee Apis mellifera (Insecta: Hymenoptera: Apidae) of New Zealand. Mitochondrial DNA. 3:66–67.
- Takahashi J, Deowanish S, Okuyama H. 2018. The complete mitochondrial genome of the dwarf honeybees, Apis florea and Apis andreniformis (Hymenoptera: Apidae) in Thailand. Mitochondrial DNA. 3:350–353.
- Takahashi J, Tingek S, Okuyama H. 2017b. The complete mitochondrial DNA sequence of endemic honeybee Apis nuluensis (Insecta: Hymenoptera: Apidae) inhabiting Mount Kinabalu in Sabah Province, Borneo Island. Mitochondrial DNA. 2:585–586.
- Takahashi J, Wakamiya T, Kiyoshi T, Uchiyama H, Yajima S, Kimura K, Nomura T. 2016. The complete mitochondrial genome of the Japanese honeybee, Apis cerana japonica (Insecta: Hymenoptera: Apidae). Mitochondrial DNA. 1:156–157.
- Takahashi J, Deowanish S, Okuyama H. 2017d. Analysis of the complete mitochondrial genome of the giant honeybee, Apis dorsata, (Hymenoptera: Apidae) in Thailand. Conserv Genet Res. doi.org/10.1007/s12686-017-0942-7
- Takahashi J, Hadisoesilo S, Okuyama H, Hepburn RH. 2017c. Analysis of the complete mitochondrial genome of Apis nigrocincta (Insecta: Hymenoptera: Apidae) on Sangihe Island in Indonesia. Conserv Genet Res.
- Takahashi J, Rai J, Wakamiya T, Okuyama H. 2017a. Characterization of the complete mitochondrial genome of the giant black Himalayan honeybee (Apis laboriosa) from Nepal. Conserv Genet Res. doi: 10.1007/s12686-017-0765-6
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wakamiya T, Tingek S, Okuyama H, Kiyoshi T, Takahashi J. 2017. The complete mitochondrial genome of the cavity-nesting honeybee, Apis koschevnikovi (Insecta: Hymenoptera: Apidae). Mitochondrial DNA. 2:24–25.
