Abstract
Bambusa ventricosa is one important ornamental bamboo. In this study, we generated the complete chloroplast (cp) genome of B. ventricosa via genome-skimming method. The complete cp genome is 139,460 base-pairs (bp) in size, with a pair of inverted repeats of 21,794 bp which separate a large single copy region of 82,996 bp and a small single copy region of 12,876 bp. The genome contains a total of 131 genes, including 84 protein-coding genes, 8 ribosomal RNAs and 39 transfer RNAs. The GC content of the cp genome is 38.9%. Moreover, phylogenomic inference robustly placed B. ventricosa in the paleotropical woody bamboos lineage.
Bambusa ventricosa McClure, commonly called buddha bamboo, is a tropical woody bamboo with important ornamental value in the family Poaceae (Bambusoideae: Bambuseae) (Li et al. Citation2006). Unfortunately, the wild populations of B. ventricosa have been dramatically fragmented and decreased over the past decades, largely due to the agricultural activities. The genetic and genomic data of this bamboo are very scarce at present (Zhou et al. Citation2017). In this study, we characterized the complete chloroplast (cp) genome of B. ventricosa using the genome-skimming sequencing approach (Zhang and Chen Citation2016), which will provide essential resource for both conservation and utilization of this economically important bamboo.
Total genomic DNA was extracted from fresh leaves of B. ventricosa grown in Bamboo Garden of Sun Yat-sen University, in Guangzhou, Guangdong province of China. The voucher specimen was deposited at the Trees Herbarium of Sun Yat-sen University (accession number ZXZ181001). Illumina paired-end library was constructed and sequenced in Beijing Genomics Institute (BGI) in Shenzhen, China. The cp genome was assembled using CLC Genomics Workbench v7.5 (CLC Bio, Aarhus, Denmark), as conducted in previous studies (Wang et al. Citation2016). The assembled cp genome was annotated using DOGMA (Wyman et al. Citation2004), coupled with manual check and adjustment.
The cp genome of B. ventricosa (GenBank accession MH410121) is 139,460 base-pairs (bp) in size with high coverage (mean 316×), showing a typical quadripartite structure: one large single copy region of 82,996 bp and one small single copy region of 12,876 bp being separated by two inverted repeats of 21,794 bp. There are a total of 131 genes in the cp genome, including 84 protein-coding genes, 8 ribosomal RNA genes, and 39 transfer RNA genes. Nine protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rps12, rps16) contain one intron each while another one gene (ycf3) had two introns. Protein-coding regions (CDS) make up 43.2% of B. ventricosa cp genome and the overall GC content of this cp genome is 38.9%. As expected, the B. ventricosa cp genome displays a highly conserved genome structure of paleotropical woody bamboos (Bambuseae) (Wu et al. Citation2009).
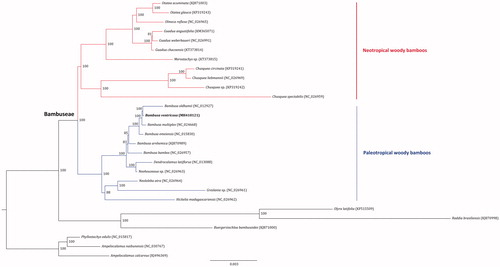
Phylogenomic analysis was performed based on the complete cp genomes of 28 bamboos using RAxML v.8.2.8 (Stamatakis Citation2014). The resulted tree showed that B. ventricosa was clustered in the clade of paleotropical woody bamboos with high support value (). The sister relationship of B. ventricosa and B. oldhamii was highly supported. The phylogenetic relationships among the sampled Bambuseae species were robustly resolved as well, in agreement with previous studies (Wang et al. Citation2018).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
Reference
- Li DZ, Wang ZP, Zhu ZD, Xia NH, Jia LZ, Guo ZH, Yang GY, Stapleton CMA. 2006. Bambuseae (Poaceae). In: Wu, ZY, Raven, PH, Hong, DY, editors. Vol. 22, Flora of China. Beijing: Science Press and Missouri Botanical Garden Press.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wang WC, Chen SY, Zhang XZ. 2016. Chloroplast genome evolution in Actinidiaceae: clpP loss, heterogenous divergence and phylogenomic practice. PLoS ONE. 11:e0162324
- Wang WC, Chen SY, Zhang XZ. 2018. Whole-genome comparison reveals divergent IR borders and mutation hotspots in chloroplast genomes of herbaceous bamboos (Bambusoideae: Olyreae). Molecules. 23:1537.
- Wu FH, Kan DP, Lee SB, Daniell H, Lee YW, Lin CC, Lin NS, Lin CS. 2009. Complete nucleotide sequence of Dendrocalamus latiflorus and Bambusa oldhamii chloroplast genomes. Tree Physiol. 29:847–856.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhang XZ, Chen SY. 2016. Genome skimming reveals the complete chloroplast genome of Ampelocalamus naibunensis (Poaceae: Bambusoideae: Arundinarieae) with phylogenomic implication. Mitochondrial DNA B Resour. 1:635–637.
- Zhou MY, Zhang YX, Haevermans T, Li DZ. 2017. Towards a complete generic-level plastid phylogeny of the paleotropical woody bamboos (Poaceae: Bambusoideae). Taxon. 66:539–553.