Abstract
Kadsura (Schisandraceae) comprises ca. 16 species and widely distributes in eastern Asia. Many species in the genus were used as important medicinal plants in China and other Asian countries. In present study, we described the first Kadsura chloroplast genome, Kadsura longipedunculata, based on illumine pair-end sequencing data. The complete plastome length is 145,346 bp long with a large single copy, a small single copy, and two inverted repeat regions of length 94,947, 17,752, and 16,323 bp, respectively. A total of 118 genes were identified, including 75 protein-coding gens, 8 rRNA genes, and 34 tRNA genes. Eighteen plastome accessions from Austrobaileyales, Nymphaeales, and Magnoliales were selected to assess the phylogenetic placement of genus.
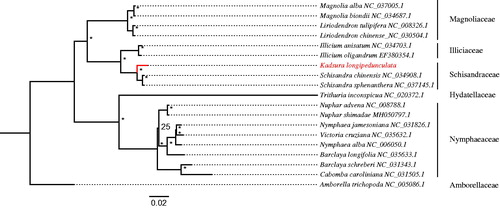
The genus Kadsura includes 16 species of scandent and twining woody vines with relatively primitive morphological characters placed in a basal angiosperm family, Schisandraceae (Austrobaileyales). Many species in the genus were used as important medicinal plants in China and other Asian countries (Zhang et al. Citation2015). Previous molecular phylogenetic study has shown that both Kadsura and its sister genus Schisandra are not monophyletic (Liu et al. Citation2006; Zhang et al. Citation2015). Kadsura longipedunculata, the Chinese Kadsura vine, is a widely used medicinal plant native to eastern China (Mulyaningsih et al. Citation2010). In this study, we assembled and characterized the complete plastome of K. longipedunculata. It is the first complete plastome reported in this genus, and will provide potential genetic resources for elucidating possible evolutionary relationships between Kadsura and Schisandra in Schisandraceae.
The plant sample collected from Xuansheng, Anhui, China (Voucher No. WH1803007255, deposited at Zhejiang Sci-Tech University). Total genomic DNA was extracted from fresh leaves of K. longipedunculata individual using DNA Plantzol Reagent (Invitrogen, Carlsbad, USA), according to the manufacturer’s instructions. Whole plastome sequences were generated using Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA, USA). In total, ca. 18.1 million high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. The CLC de novo assembler (CLC Bio, Aarhus, Denmark), BLAST, GeSeq (Tillich et al. Citation2017), and tRNAscan-SE v1.3.1 were used to align, assemble, and annotate the plastome (Schattner et al. Citation2005).
The full length of K. longipedunculata chloroplast genome (GenBank acession MH550622) was 145,346 bp and consist of a large single copy region (LSC with 94,947 bp), a small single copy region (SSC with 17,752 bp), and a pair of inverted repeat regions (IRs) of 16,323 bp. The overall GC content of the K. longipedunculata cp genome was 39.7% and the GC content in the LSC, SSC, and IR regions are 38.6%, 35%, and 45.6%, respectively. A total of 118 genes were contained in the cp genome (75 protein-coding genes, 8 rRNA genes, and 34 tRNA genes). Twelve genes had two copies, which included 2 PCG genes (ndhb, rps7), 6 tRNA genes (trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and all 4 rRNA species (rrn4.5, rrn5, rrn16 and rrn23). Among the protein-coding genes, two genes (clpP and ycf3) contained two introns, and other seven genes (atpF, ndhA, ndhB, rpl2, rpoC1, rps16) had one intron each.
For conducting its phylogenetic relationships with present reported complete chloroplast genomes, eighteen chloroplast genomes sequences from the order of Austrobaileyales, Nymphaeales, and Magnoliales were selected (). All of genomes were fully aligned with MAFFT v7.3 (Katoh and Standley Citation2013), and the maximum-likelihood (ML) inference was performed using GTR model with support for branches evaluated by 1000 bootstrap replicates with RAxML v.8.2.1 (Stamatakis Citation2014) on the CIPRES cluster service (Miller et al. Citation2010). We hope the complete plastome of K. longipedunculata will provide a valuable resource for further genetic conservation, phylogenomic, and evolution studies in the genus and family.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Liu Z, Hao G, Luo YL, Thien BS, Rosso W, Lu A, Chen ZD. 2006. Phylogeny and androecial evolution in Schisandraceae, inferred from sequences of nuclear ribosomal DNA ITS and chloroplast DNA trnL‐F regions. Inter J Plant Sci. 167:539–550.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. New Orleans, LA: Proceedings of the gateway computing environments workshop (GCE).
- Mulyaningsih S, Youns M, El-Readi MZ, Ashour ML, Nibret E, Sporer F, Herrmann F, Reichling J, Wink M. 2010. Biological activity of the essential oil of Kadsura longipedunculata (Schisandraceae) and its major components. J Pharm Pharmacol. 62:1037–1044.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Zhang J, Chen M, Dong X, Lin R, Fan J, Chen ZD. 2015. Evaluation of four commonly used DNA barcoding loci for Chinese medicinal plants of the family Schisandraceae. Plos One. 10:e0125574