Abstract
Atraphaxis jrtyschensis (Polygonacae) is an endangered desert shrub endemic to China in Xinjiang province with great ecological importance for sand fixation. However, its genomic resources are still very limited. Here, we generated the first chloroplast (cp) genome of A. jrtyschensis using genome skimming sequencing. The whole cp genome is 164,192 bp and comprises 130 genes, including 83 protein-coding genes, 37 tRNA genes, 8 rRNA genes, and 2 pseudogenes (rpl23). The overall GC content of A. jrtyschensis cp genome is 37.5%. The phylogenic analysis placed A. jrtyschensis at the base of Trib Rumiceae, which contained the genera Rheum and Oxyria. This study will be useful for future researches to investigate the conservation genetics and potential applications in sand fixation of the endangered desert shrub.
Atraphaxis jrtyschensis Yang et Han (Polygonaceae) is an endangered desert shrub, which is endemic in the Buerjin County in Xinjiang province of Northwest China (Feng et al. Citation2003). It is an ecologically and economically important shrub for sand fixation and forage. However, the wild resources of A. jrtyschensis have been decreased dramatically and only few populations can be found in the field. It is now necessary to take effective methods to conserve this desert shrub. As far as we know, there is no study that has been focused on this shrub except a phylogenetic study of trib Atraphaxideae based on cp DNA fragments (Sun and Zhang Citation2012). Comprehensive genomic resources are absent for A. jrtyschensis, which limits the understanding of its conservation genetics at the genome level. In this study, we sequenced, assembled, and annotated the complete cp genome of A. jrtyschensis based on the sequencing data.
The fresh leaves of A. jrtyschensis were collected from the Buerjin County (86.867694E, 47.695674 N) and vouchers were deposited in Xinjiang University (Accession Number ML0025). Total genomic DNA were extracted with the CTAB method (Doyle and Doyle Citation1987). The complete-genome sequencing was conducted with 150 bp pair-end (PE) reads on the Illumina Hiseq Platform(Illumina, San Diego, CA). In total, 5.0 GB raw data was obtained and approximately 10 million high quality clean reads were collected after filtering low-quality reads. The complete circular cp genome were assembled using the softwere Velvet (Zerbino and Birney Citation2008). After manual adjustment, we performed an automated genome annotation with a recently developed command-line application called Plastome Annotator (Plann) (Huang and Cronk Citation2015). Then, we got a physical map using the software OGDRAW (Lohse et al. Citation2013). A maximum likelihood (ML) tree was reconstructed using the software RaxML version 8 (Stamatakis Citation2014) from the alignments created by the MEGA6.06 (Tamura et al. Citation2013). Finally, the complete chloroplast genome sequence and its annotations were submitted to GenBank with an accession number of MG878984.
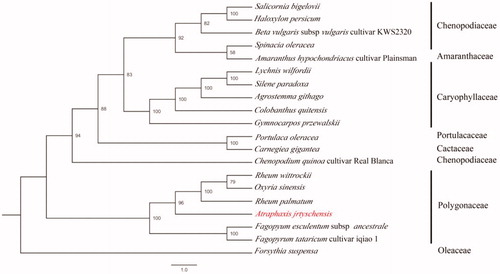
The chloroplast genome of A. jrtyschensis showed a typical quadripartite structure with a length of 164,192 bp, which contained inverted repeats (IR) of 24,152 bp separated by a large single-copy (LSC) and a small single copy (SSC) of 88,878 bp and 27,010 bp, respectively. The cpDNA contains 130 genes, comprising 83 protein-coding genes, 37 tRNA genes, 8 rRNA genes, and 2 pseudogenes (rpl23). Among the annotated genes, 10 of them contained only 1 intron (rps12, trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnV-UAC, petB, petD, and ndhA), and 5 genes (trnA-UGC, trnI-GAU, rpl2, ndhB, and clpP) contained 2 introns. The overall GC content of the plastome is 37.5%, while the corresponding values of the LSC, SSC, and IR regions are 35.5, 33.0, and 43.6%, respectively. Further, the phylogenetic analysis of 20 plastid genomes with Forsythia suspensa used as the outgroup showed that A. jrtyschensis is related to the Trib Rumiceae, which includes the genera Rheum and Oxyria (). The A. jrtyschensis genome will be a valuable genetic resource and can be used for population genetic and phylogenetic studies of A. jrtyschensis.
Figure 1. The phylogenetic tree based on seven complete chloroplast genome sequences. Accession numbers: Rheum palmatum (KR816224), Rheum wittrockii (KY985269), Oxyria sinensis (KX774248), Fagopyrum tataricum (KX085498), Faopyrum esculentum (EU254477), Forsythia suspense (MF579702), Chenopodium quinoa (CM008430), Haloxylon persicum (NC027669), Spinacia oleracea (AJ400848), Beta vulgaris (KR230391), Amaranthus hypochondriacus (NC030770), Salicornia bigelovii (NC027226), Portulaca oleracea (NC036236), Carnegiea gigantean (NC027618), Colobanthus quitensis (NC028080), Silene paradoxa (NC023360), Agrostemma githago (NC023357), Gymnocarpos przewalskii (NC036812), Lychnis wilfordii (NC035225).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Feng Y, Yan C, Yin LK. 2003. The endemic species and distribution in Xinjiang. Acta Bot Boreali-Occidentalia Sin. 23:263–273.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:w575–w581.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Sun YX, Zhang ML. 2012. Molecular phylogeny of tribe Atraphaxideae (Polygonaceae) evidenced from five cpDNA genes. J Arid Land. 4:180–190.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
