Abstract
To understand the evolution of the swimming crab Thalamita crenata, the complete mitochondrial genome of T. crenata from China was sequenced and analyzed. The circular mitogenome sequence was 15,787 bp in length, made up of 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes and a control region. The overall mitogenome composition was 34.40% for A, 11.55% for G, 35.31% for T, and 18.74% for C, respectively, with a high A + T content of 69.71%. Phylogenetic analysis showed that T. crenata was closest to the genus Charybdis.
The spiny rock crab, Thalamita crenata, under the Family Portunidae is mainly distributed in Western Pacific and Indian Ocean (Dai et al. Citation1986; Dai and Yang Citation1991; Stephenson Citation1972). It inhabits the rocky and muddy intertidal regions of mangrove forests (Vezzosi et al. 1994). Unlike other big-size economic portunids, such as the mud crab Scylla paramamosain and the red crab Charybdis feriatus, T. crenata has less economic value (Manickaraja and Balasubramanian Citation2009). However, its natural population is under threat from local fishing pressure (Manickaraja and Balasubramanian Citation2009). In order to gain a deep understanding of the phylogeny and species evolution, we described, in this study, the complete mitochondrial genome of T. crenata from China.
Specimens of T. crenata were collected from Weizhou Island (21.0234°N, 109.0940°E), Guangxi province, China and preserved in pure ethanol in Marine Biology Institute of Shantou University. Total genomic DNA was extracted from muscle tissues and the complete mitogenome sequence was obtained by long and conventional PCR. Additionally, the complete mitochondrial genomes of 17 brachyuran species and one shrimp (Hapiosquilla harpax) were downloaded from NCBI. The ND6 gene showed a high degree of heterogeneity and caused poor phylogenetic performance (Miya and Nishida Citation2000), so it was omitted in next analysis. The maximum-likelihood (ML) phylogenetic tree was constructed by MEGA7 software based on 12 protein-coding genes.
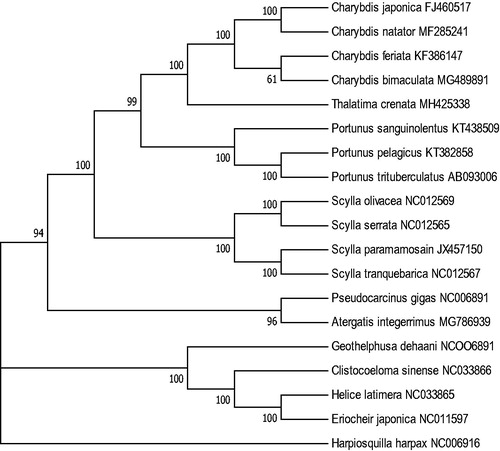
The complete mitochondrial genome of T. crenata was 15,787 bp in length (GenBank accession number: MH425338), including 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and a control region. The overall mitogenome composition was 34.40% for A, 11.55% for G, 35.31% for T, and 18.74% for C, respectively, with a high A + T content of 69.71%. The phylogenetic analysis showed that T. crenata was closest to the genus Charybdis (). The control region (D-loop) was 898 bp long and between the genes tRNAPro and tRNAPhe, with a a high similarity of 95% to the corresponding region of T. crenata collected from Australia (Tan et al. Citation2016).
Disclosure statement
The authors declare no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Dai AY, Yang SL, Song YZ. 1986. Marine crabs in China Sea. Beijing. Marine Publishing Company; p.194–196.
- Dai A, Yang SL. 1991. Crabs of the China seas. Beijing: China Ocean Press; p.1–74.
- Manickaraja M, Balasubramanian TS. 2009. Occurrence of the deep sea crab, Thalamita crenata in shallow water gillnet (mural valai) operation at Tharuvaikulam, north of Tuticorin. Mar Fish Inf Serv Tech Ext Ser. 201:28.
- Miya M, Nishida M. 2000. Use of mitogenomic information in teleostean molecular phylogenetics: a tree-based exploration under the maximum-parsimony optimality criterion. Mol Phylogenet Evol. 17:437–455.
- Stephenson W. 1972. Portunid crabs from the ludo-West Pacific and Western America in the zoologica. Museum Copenhagen (Decapoda, Brachyura, Portunidae). Steenstrupia. 2:127–156.
- Tan MH, Gan HM, Lee YP, Austin CM. 2016. The complete mitogenome of the swimming crab Thalamita crenata (Rüppell, 1830) (Crustacea; Decapoda; Portunidae). Mitochondr DNA. 27:1275–1272.
- Vezzosi R, Barbaresi S, Anyona D, Vannini M. 1994. Activity patterns in Thalamita crenata Portunidae, decapoda: a shaping by the tidal cycles. Mar Behav Physiol. 24:207–214.